Welcome to China Oncology,
China Oncology ›› 2022, Vol. 32 ›› Issue (10): 979-989.doi: 10.19401/j.cnki.1007-3639.2022.10.006
• Article • Previous Articles Next Articles
ZHU Yi1,3( ), XIAO Bin1, LIU Jiahui2, HUANG Ling2, SUN Zhaohui2, LI Linhai1
), XIAO Bin1, LIU Jiahui2, HUANG Ling2, SUN Zhaohui2, LI Linhai1
Received:2022-03-24
Revised:2022-08-04
Online:2022-10-30
Published:2022-11-29
Share article
CLC Number:
ZHU Yi, XIAO Bin, LIU Jiahui, HUANG Ling, SUN Zhaohui, LI Linhai. Expression, localization, biological role and proteomics study of circ-0003910 in HER2-positive breast cancer[J]. China Oncology, 2022, 32(10): 979-989.
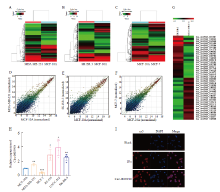
Fig. 2
Circ-0003910 was up-regulated in HER2 positive breast cancer cells A-C: Heat map of differentially expressed circRNA; D-F: Scatter plots of differentially expressed circRNA; G: Heat map of the top 20 circRNA with the highest differential expression; H: Expression of circ-0003910 in five types of breast cancer cells and normal breast cells; I: Subcellular localization of circ-0003910 in SK-BR-3 cells; *: P<0.05, **: P<0.01, ***: P<0.001, compared with MCF-10A."
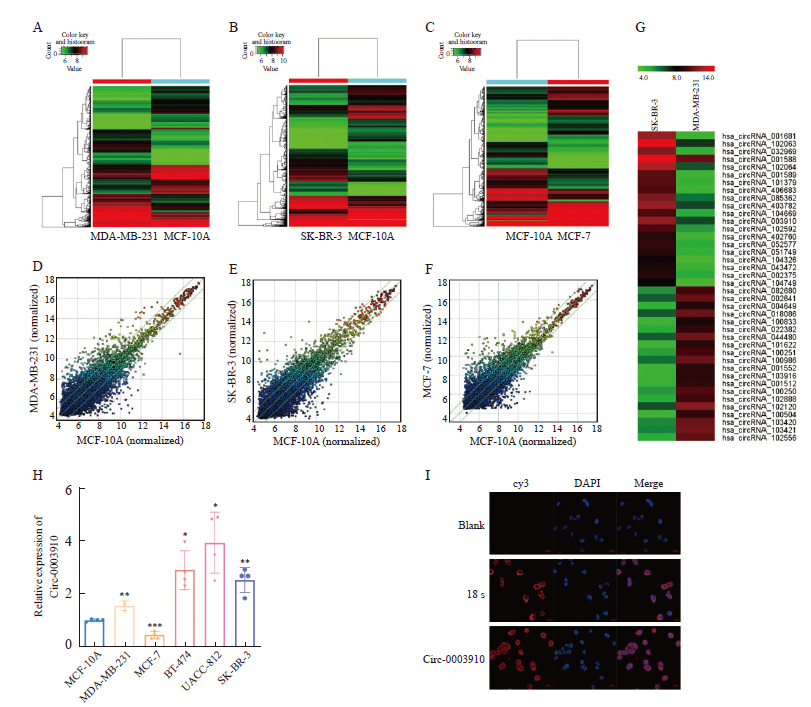
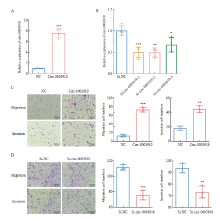
Fig. 4
Circ-0003910 promotes breast cancer cell migration and invasion in vitro A-B: The mRNA expression level of CIRC-0003910 was detected after overexpression and knockdown of circ-0003910 by RTFQ-PCR; C: Transwell assay detected the migration and invasion of MDA-MB-231 cells when circ-0003910 was overexpressed; D: Transwell assay detects the migration and invasion of SK-BR-3 cells when circ-0003910 was knocked down. **: P<0.01, compared with each other; ***: P<0.001, compared with each other."

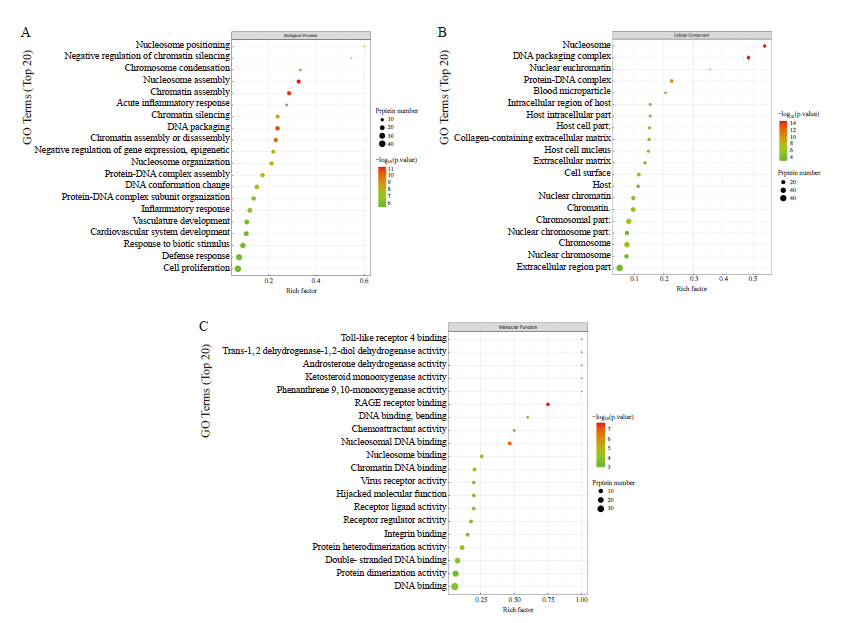
Fig. 5
GO enrichment analysis of downstream differentially expressed proteins affected by circ-0003910 overexpression A: GO analysis of differentially expressed proteins enriched in biological process; B: GO analysis of differentially expressed proteins enriched in cellular component; C: GO analysis of differentially expressed proteins enriched in molecular function"
| [1] |
SUNG H, FERLAY J, SIEGEL R L, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71(3): 209-249.
doi: 10.3322/caac.21660 |
| [2] |
YEO S K, GUAN J L. Breast cancer: multiple subtypes within a tumor?[J]. Trends Cancer, 2017, 3(11): 753-760.
doi: S2405-8033(17)30175-9 pmid: 29120751 |
| [3] |
HARBECK N, GNANT M. Breast cancer[J]. Lancet, 2017, 389(10074): 1134-1150.
doi: S0140-6736(16)31891-8 pmid: 27865536 |
| [4] | 王笑. HER2阳性乳腺癌中KIF3B表达意义及其与上皮间质转化的相关性研究[D]. 青岛: 青岛大学, 2021. |
| WANG X. Significance of KIF3B expression in HER2 positive breast cancer and its correlation with epithelial mesenchymal transition[D] Qingdao: Qingdao University, 2021. | |
| [5] |
ZHENG Q P, BAO C Y, GUO W J, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs[J]. Nat Commun, 2016, 7: 11215.
doi: 10.1038/ncomms11215 pmid: 27050392 |
| [6] |
YIN Y T, LONG J L, HE Q L, et al. Emerging roles of circRNA in formation and progression of cancer[J]. J Cancer, 2019, 10(21): 5015-5021.
doi: 10.7150/jca.30828 pmid: 31602252 |
| [7] |
LIN X S, HUANG C, CHEN Z A, et al. CircRNA_100876 is upregulated in gastric cancer (GC) and promotes the GC cells' growth, migration and invasion via miR-665/YAP1 signaling[J]. Front Genet, 2020, 11: 546275.
doi: 10.3389/fgene.2020.546275 |
| [8] |
LIU H L, LAN T, LI H, et al. Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR[J]. Theranostics, 2021, 11(3): 1396-1411.
doi: 10.7150/thno.53227 pmid: 33391541 |
| [9] |
CHEN D S, MA W, KE Z Y, et al. CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression[J]. Cell Cycle, 2018, 17(16): 2080-2090.
doi: 10.1080/15384101.2018.1515553 pmid: 30176158 |
| [10] |
LIU Z H, ZHOU Y, LIANG G H, et al. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p[J]. Cell Death Dis, 2019, 10(2): 55.
doi: 10.1038/s41419-018-1287-1 pmid: 30670688 |
| [11] | 王晓松, 陈俊霞. 环状RNA hsa_circ_0050900在乳腺癌中的表达及其对细胞生物学行为影响的机制研究[J]. 中国癌症杂志, 2020, 30(12): 977-983. |
| WANG X S, CHEN J X. Expression of circular RNA has_circ_0050900 in breast cancer and its effect on cell function[J]. China Oncol, 2020, 30(12): 977-983. | |
| [12] |
LUO Z, RONG Z Y, ZHANG J M, et al. Circular RNA circCCDC9 acts as a miR-6792-3p sponge to suppress the progression of gastric cancer through regulating CAV1 expression[J]. Mol Cancer, 2020, 19(1): 86.
doi: 10.1186/s12943-020-01203-8 pmid: 32386516 |
| [13] |
XU J Z, SHAO C C, WANG X J, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis[J]. Cell Death Dis, 2019, 10(3): 175.
doi: 10.1038/s41419-019-1382-y |
| [14] | ZEESHAN R, MUTAHIR Z. Cancer metastasis-tricks of the trade[J]. Bosn J Basic Med Sci, 2017, 17(3): 172-182. |
| [15] |
LI T, MELLO-THOMS C, BRENNAN P C. Descriptive epidemiology of breast cancer in China: incidence, mortality, survival and prevalence[J]. Breast Cancer Res Treat, 2016, 159(3): 395-406.
doi: 10.1007/s10549-016-3947-0 |
| [16] |
ZANG J K, LU D, XU A D. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function[J]. J Neurosci Res, 2020, 98(1): 87-97.
doi: 10.1002/jnr.24356 pmid: 30575990 |
| [17] |
KRISTENSEN L S, ANDERSEN M S, STAGSTED L V W, et al. The biogenesis, biology and characterization of circular RNAs[J]. Nat Rev Genet, 2019, 20(11): 675-691.
doi: 10.1038/s41576-019-0158-7 pmid: 31395983 |
| [18] | KLINGE C M. Non-coding RNAs in breast cancer: Intracellular and intercellular communication[J]. Noncoding RNA, 2018, 4(4): 40. |
| [19] |
KIRBY E, TSE W H, PATEL D, et al. First steps in the development of a liquid biopsy in situ hybridization protocol to determine circular RNA biomarkers in rat biofluids[J]. Pediatr Surg Int, 2019, 35(12): 1329-1338.
doi: 10.1007/s00383-019-04558-2 |
| [20] |
HUANG A Q, ZHENG H X, WU Z Y, et al. Circular RNA-protein interactions: functions, mechanisms, and identification[J]. Theranostics, 2020, 10(8): 3503-3517.
doi: 10.7150/thno.42174 pmid: 32206104 |
| [21] |
HU C J, NI Z H, LI B S, et al. hTERT promotes the invasion of gastric cancer cells by enhancing FOXO3a ubiquitination and subsequent ITGB1 upregulation[J]. Gut, 2017, 66(1): 31-42.
doi: 10.1136/gutjnl-2015-309322 pmid: 26370108 |
| [22] |
REN L L, MO W J, WANG L L, et al. Matrine suppresses breast cancer metastasis by targeting ITGB1 and inhibiting epithelial-to-mesenchymal transition[J]. Exp Ther Med, 2020, 19(1): 367-374.
doi: 10.3892/etm.2019.8207 pmid: 31853313 |
| [23] |
TAFTAF R, LIU X, SINGH S, et al. ICAM1 initiates CTC cluster formation and trans-endothelial migration in lung metastasis of breast cancer[J]. Nat Commun, 2021, 12(1): 4867.
doi: 10.1038/s41467-021-25189-z pmid: 34381029 |
| [24] |
FIGENSCHAU S L, KNUTSEN E, URBAROVA I, et al. ICAM1 expression is induced by proinflammatory cytokines and associated with TLS formation in aggressive breast cancer subtypes[J]. Sci Rep, 2018, 8(1): 11720.
doi: 10.1038/s41598-018-29604-2 pmid: 30082828 |
| [25] |
ALTEVOGT P, DOBERSTEIN K, FOGEL M. L1CAM in human cancer[J]. Int J Cancer, 2016, 138(7): 1565-1576.
doi: 10.1002/ijc.29658 pmid: 26111503 |
| [26] |
LI Y, GALILEO D S. Soluble L1CAM promotes breast cancer cell adhesion and migration in vitro, but not invasion[J]. Cancer Cell Int, 2010, 10: 34.
doi: 10.1186/1475-2867-10-34 |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
沪ICP备12009617
Powered by Beijing Magtech Co. Ltd
