Welcome to China Oncology,
China Oncology ›› 2024, Vol. 34 ›› Issue (6): 537-547.doi: 10.19401/j.cnki.1007-3639.2024.06.002
• Article • Previous Articles Next Articles
DONG Jianqiao1( ), LI Kunyan1, LI Jing2, WANG Bin2, WANG Yanhong3, JIA Hongyan2(
), LI Kunyan1, LI Jing2, WANG Bin2, WANG Yanhong3, JIA Hongyan2( )(
)( )
)
Received:2024-03-07
Revised:2024-06-07
Online:2024-06-30
Published:2024-07-16
Share article
CLC Number:
DONG Jianqiao, LI Kunyan, LI Jing, WANG Bin, WANG Yanhong, JIA Hongyan. A study on mechanism of SIRT3 inducing endocrine drug resistance in breast cancer via deacetylating YME1L1[J]. China Oncology, 2024, 34(6): 537-547.
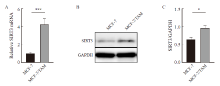
Fig. 2
SIRT3 is highly expressed in tamoxifen-resistant breast cancer. A: SIRT3 mRNA expression was detected by RTFQ-PCR. B, C: SIRT3 protein expression and quantification in both cell lines were detected by Western blot. *: P<0.05, compared with each other; ***: P<0.001, compared with each other."
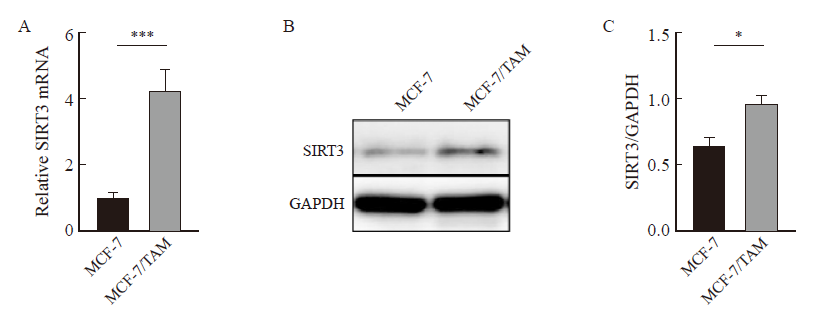
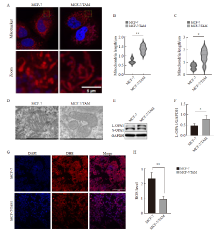
Fig. 3
Detection of mitochondrial morphology and function in breast cancer after endocrine resistance A, B: Mitochondrial length was observed and quantified by fluorescence staining (scale bar=5 μm); C, D: Mitochondrial morphology was observed and quantified by transmission electron microscopy (scale bar=200 nm); E, F: Western blot was used to detect and quantify L-OPA1 protein in parental cells and drug-resistant cells. G, H: DHE staining assessment, ROS levels and quantification in parental and resistant cells (scale bar=200 μm). *: P<0.05, compared with each other; **: P<0.01, compared with each other."
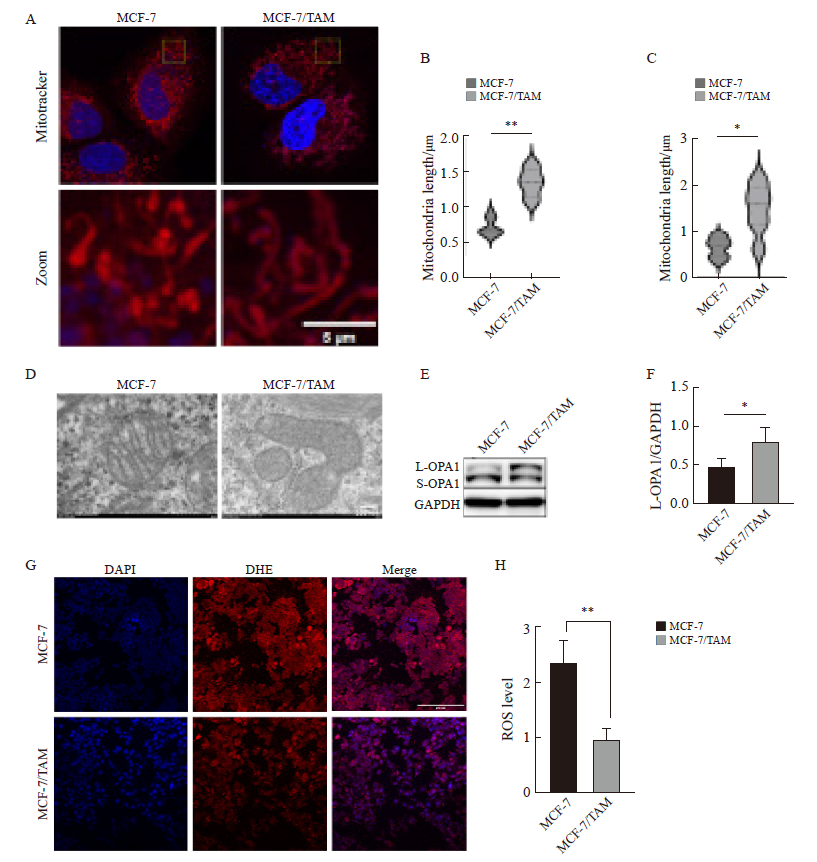
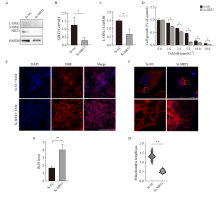
Fig. 4
Cell proliferation, mitochondrial morphology, and function were examined by SIRT3 knockdown in drug-resistant cells A, B, C: Western blot was used to detect the expression and quantification of SIRT3 protein and L-OPA1 protein after SIRT3 knockdown. D: CCK-8 assay was used to detect the cell viability under tamoxifen treatment; E, G: DHE probe was used to detect the intracellular ROS level and quantification after SIRT3 knockdown (scale bar=200 μm); F, H: Immunofluorescence detection of mitochondrial length changes and quantification after SIRT3 knockdown (scale bar=5 μm). *: P<0.05, Si-NC compared with Si-SIRT3."
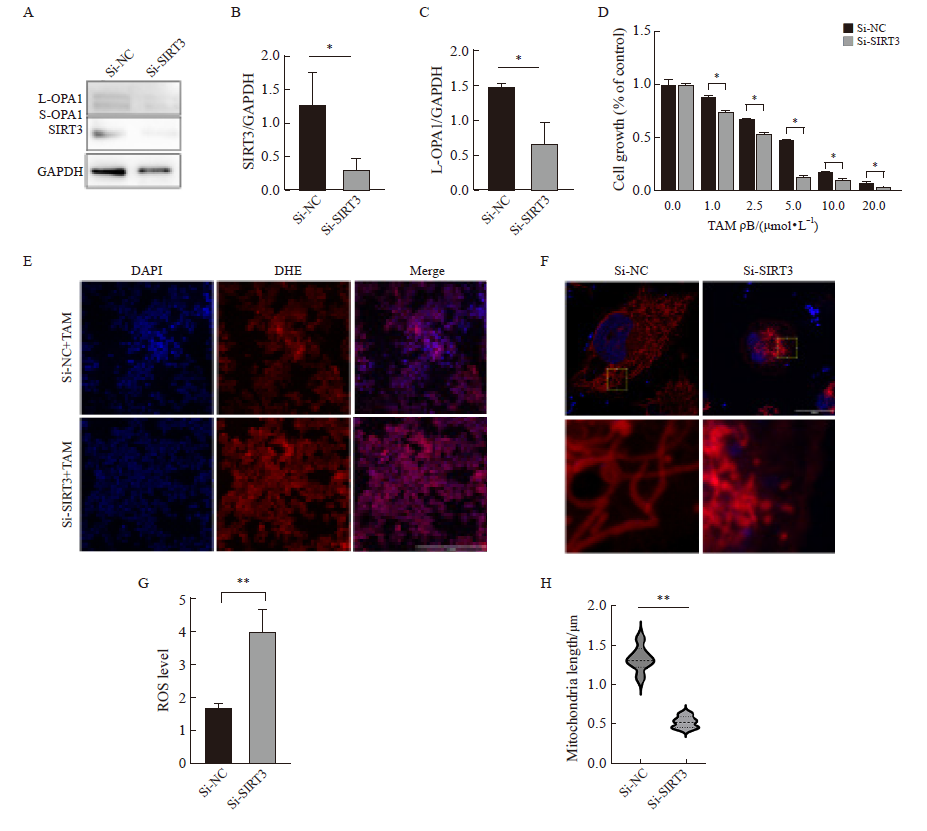

Fig. 5
Overexpression of SIRT3 in parental cells examined cell proliferation, mitochondrial morphology, and function A, B, C: Western blot was used to detect the expression and quantification of L-OPA1 protein after SIRT3 overexpression; D: After SIRT3 was overexpressed in parental cells, CCK-8 assay was used to detect the cell viability under tamoxifen treatment; E, G: Detection and quantification of mitochondrial membrane potential by JC-1 probe (scale bar=200 μm); H, I: Fluorescence staining for mitochondrial morphology and quantification (scale bar=5 μm). *: P<0.05, compared with each other; **: P<0.01, compared with each other; ***: P<0.001, compared with OE-SIRT3 group."
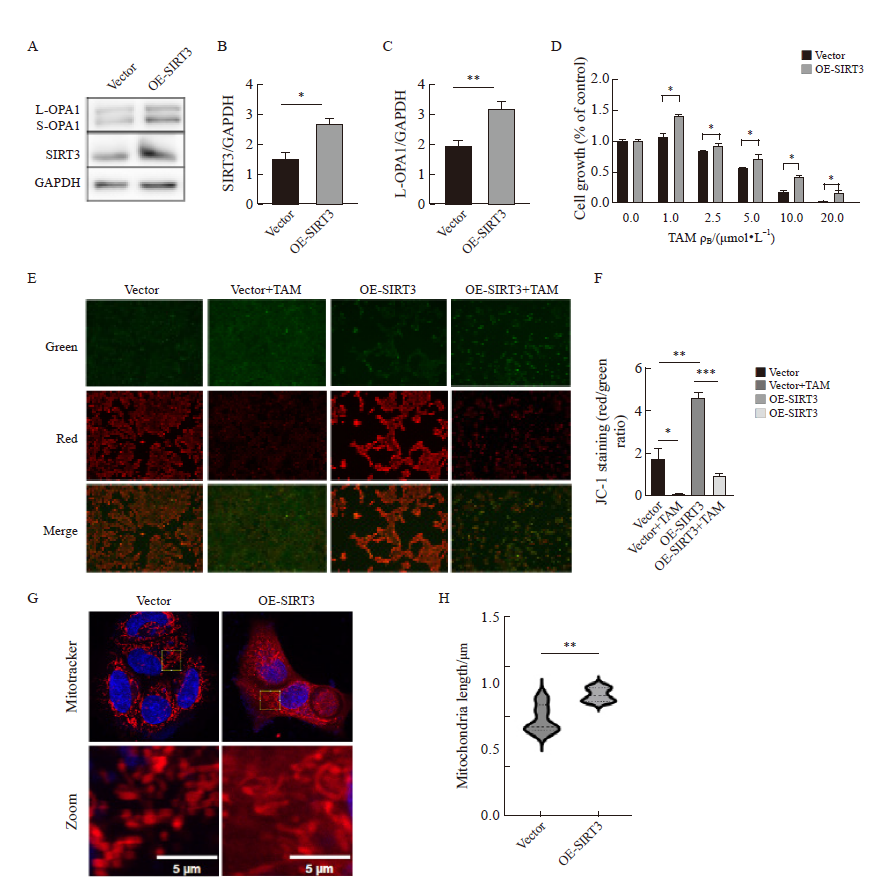
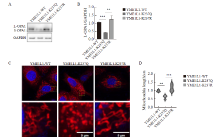
Fig. 6
Changes in mitochondrial morphology caused by overexpression of YME1L1 in parental cells A, B: YME1L1 was overexpressed, and the expression and quantification of L-OPA1 protein were detected by Western blot. C, D: Length and quantification of mitochondria by fluorescent staining (scale bar=5 μm). **: P<0.01, compared with each other; ***: P<0.001, compared with YME1L1-K237Q."
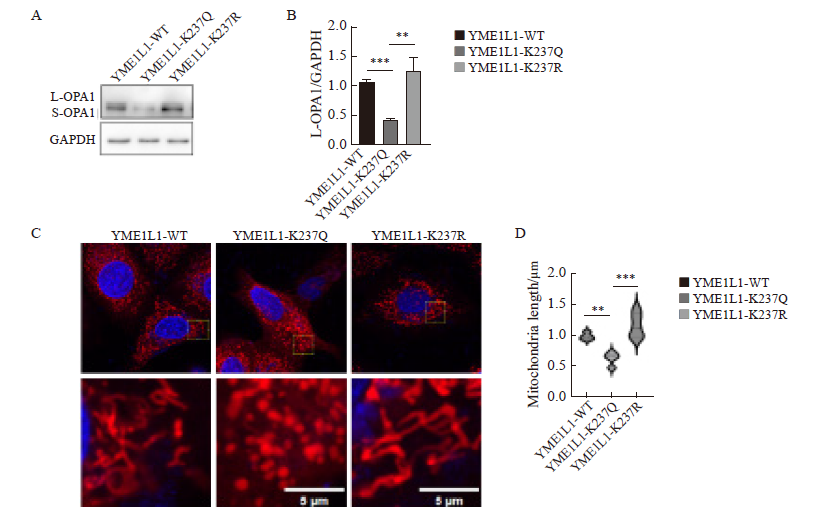

Fig. 7
SIRT3 regulates the acetylation status of YME1L1 A-G: IP was used to detect the interaction between YME1L1 and SIRT3 and quantify. *: P<0.05, compared with each other; **: P<0.01, compared with each other; ***: P<0.001, compared with each other. H: Co-localization of YME1L1 and SIRT3 was detected by immunofluorescence (scale bar=50 μm)."
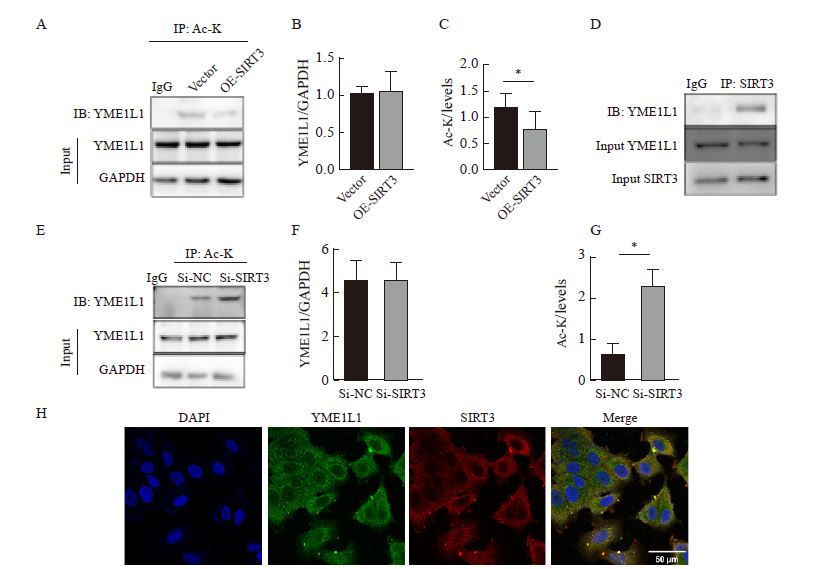
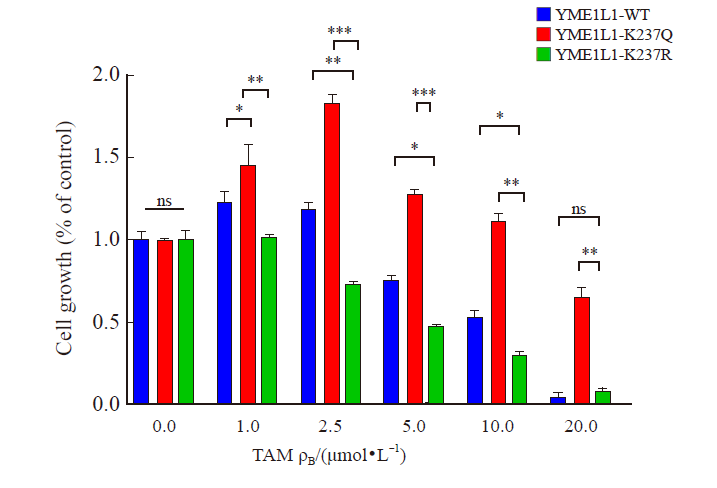
Fig. 8
Deacetylating Y ME1L1 promotes tamoxifen resistance in breast cancer CCK-8 assay was used to detect cell viability under tamoxifen treatment. *: P<0.05, compared with each other; **: P<0.01, compared with each other; ***: P<0.001, compared with each other. ns: No significance, compared with YME1L1-WT or YME1L1-K237R."
| [1] | BRAY F, LAVERSANNE M, SUNG H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA A Cancer J Clin, 2024, 74(3): 229-263. |
| [2] | CAO W, CHEN H D, YU Y W, et al. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020[J]. Chin Med J, 2021, 134(7): 783-791. |
| [3] | SIEGEL R L, GIAQUINTO A N, JEMAL A. Cancer statistics, 2024[J]. CA Cancer J Clin, 2024, 74(1): 12-49. |
| [4] | VOUDOURI K, BERDIAKI A, TZARDI M, et al. Insulin-like growth factor and epidermal growth factor signaling in breast cancer cell growth: focus on endocrine resistant disease[J]. Anal Cell Pathol, 2015, 2015: 975495. |
| [5] | ZAHEDIPOUR F, JAMIALAHMADI K, KARIMI G. The role of noncoding RNAs and sirtuins in cancer drug resistance[J]. Eur J Pharmacol, 2020, 877: 173094. |
| [6] | LIU F, YUAN L H, LI L, et al. S-sulfhydration of SIRT3 combats BMSC senescence and ameliorates osteoporosis via stabilizing heterochromatic and mitochondrial homeostasis[J]. Pharmacol Res, 2023, 192: 106788. |
| [7] |
ZHANG J, XIANG H G, LIU J, et al. Mitochondrial Sirtuin 3: new emerging biological function and therapeutic target[J]. Theranostics, 2020, 10(18): 8315-8342.
doi: 10.7150/thno.45922 pmid: 32724473 |
| [8] |
ZHANG L, REN X C, CHENG Y, et al. Identification of Sirtuin 3, a mitochondrial protein deacetylase, as a new contributor to tamoxifen resistance in breast cancer cells[J]. Biochem Pharmacol, 2013, 86(6): 726-733.
doi: 10.1016/j.bcp.2013.06.032 pmid: 23856293 |
| [9] | HUBER-KEENER K J, LIU X P, WANG Z, et al. Differential gene expression in tamoxifen-resistant breast cancer cells revealed by a new analytical model of RNA-Seq data[J]. PLoS One, 2012, 7(7): e41333. |
| [10] | Disturbance of mitochondrial dynamics and mitophagy in sepsis-induced acute kidney injury-PubMed[Internet]. [cited 2024 Mar 2]. https://pubmed.ncbi.nlm.nih.gov/31479679/. |
| [11] |
BAEK M L, LEE J, PENDLETON K E, et al. Mitochondrial structure and function adaptation in residual triple negative breast cancer cells surviving chemotherapy treatment[J]. Oncogene, 2023, 42(14): 1117-1131.
doi: 10.1038/s41388-023-02596-8 pmid: 36813854 |
| [12] | ADEBAYO M, SINGH S, SINGH A P, et al. Mitochondrial fusion and fission: the fine-tune balance for cellular homeostasis[J]. FASEB J, 2021, 35(6): e21620. |
| [13] | RUAN Y, LI H, ZHANG K, et al. Loss of Yme1L perturbates mitochondrial dynamics[J]. Cell Death Dis, 2013, 4(10): e896. |
| [14] | ZAMBERLAN M, BOECKX A, MULLER F, et al. Inhibition of the mitochondrial protein Opa1 curtails breast cancer growth[J]. J Exp Clin Cancer Res, 2022, 41(1): 95. |
| [15] |
HANKER A B, SUDHAN D R, ARTEAGA C L. Overcoming endocrine resistance in breast cancer[J]. Cancer Cell, 2020, 37(4): 496-513.
doi: S1535-6108(20)30149-5 pmid: 32289273 |
| [16] |
CARRICO C, MEYER J G, HE W J, et al. The mitochondrial acylome emerges: proteomics, regulation by sirtuins, and metabolic and disease implications[J]. Cell Metab, 2018, 27(3): 497-512.
doi: S1550-4131(18)30068-8 pmid: 29514063 |
| [17] | LIU L G, LI Y, CAO D Y, et al. SIRT3 inhibits gallbladder cancer by induction of AKT-dependent ferroptosis and blockade of epithelial-mesenchymal transition[J]. Cancer Lett, 2021, 510: 93-104. |
| [18] | S O, Q Z, L L, K Z, Z L, P L, et al. The double-edged sword of SIRT3 in cancer and its therapeutic applications[J]. Front Pharmacol, 2022, 13: 871560. |
| [19] | KENNY T C, HART P, RAGAZZI M, et al. Selected mitochondrial DNA landscapes activate the SIRT3 axis of the UPRmt to promote metastasis[J]. Oncogene, 2017, 36(31): 4393-4404. |
| [20] | ASHRAF N, ZINO S, MACINTYRE A, et al. Altered sirtuin expression is associated with node-positive breast cancer[J]. Br J Cancer, 2006, 95(8): 1056-1061. |
| [21] |
CHEN S Y, YANG X, YU M, et al. SIRT3 regulates cancer cell proliferation through deacetylation of PYCR1 in proline metabolism[J]. Neoplasia, 2019, 21(7): 665-675.
doi: S1476-5586(18)30666-3 pmid: 31108370 |
| [22] | KIM H S, PATEL K, MULDOON-JACOBS K, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress[J]. Cancer Cell, 2010, 17(1): 41-52. |
| [23] |
DESOUKI M M, DOUBINSKAIA I, GIUS D, et al. Decreased mitochondrial SIRT3 expression is a potential molecular biomarker associated with poor outcome in breast cancer[J]. Hum Pathol, 2014, 45(5): 1071-1077.
doi: 10.1016/j.humpath.2014.01.004 pmid: 24746213 |
| [24] |
FINLEY L W S, CARRACEDO A, LEE J, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization[J]. Cancer Cell, 2011, 19(3): 416-428.
doi: 10.1016/j.ccr.2011.02.014 pmid: 21397863 |
| [25] | ANDREWS R M, KUBACKA I, CHINNERY P F, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA[J]. Nat Genet, 1999, 23(2): 147. |
| [26] | OHBA Y, MACVICAR T, LANGER T. Regulation of mitochondrial plasticity by the i-AAA protease YME1L[J]. Biol Chem, 2020, 401(6/7): 877-890. |
| [27] |
ANAND R, WAI T, BAKER M J, et al. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission[J]. J Cell Biol, 2014, 204(6): 919-929.
doi: 10.1083/jcb.201308006 pmid: 24616225 |
| [28] |
HERKENNE S, EK O, ZAMBERLAN M, et al. Developmental and tumor angiogenesis requires the mitochondria-shaping protein Opa1[J]. Cell Metab, 2020, 31(5): 987-1003.e8.
doi: S1550-4131(20)30189-3 pmid: 32315597 |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
沪ICP备12009617
Powered by Beijing Magtech Co. Ltd
