Welcome to China Oncology,
China Oncology ›› 2022, Vol. 32 ›› Issue (4): 324-334.doi: 10.19401/j.cnki.1007-3639.2022.04.005
• Article • Previous Articles Next Articles
WANG Chuntao1( )(
)( ), GE Anxing1, WU Hongyan1, ZHANG Xueyan1, YANG Sheng2, YUAN Hongxiang3, CHENG Yanping2, FENG Yanlu2, LU Xinyuan2, LIANG Geyu2(
), GE Anxing1, WU Hongyan1, ZHANG Xueyan1, YANG Sheng2, YUAN Hongxiang3, CHENG Yanping2, FENG Yanlu2, LU Xinyuan2, LIANG Geyu2( )(
)( )
)
Received:2021-08-18
Revised:2022-03-02
Online:2022-04-30
Published:2022-05-07
Contact:
LIANG Geyu
E-mail:yammy_taotao@163.com;gyliang@seu.edu.cn
Share article
CLC Number:
WANG Chuntao, GE Anxing, WU Hongyan, ZHANG Xueyan, YANG Sheng, YUAN Hongxiang, CHENG Yanping, FENG Yanlu, LU Xinyuan, LIANG Geyu. The association between cervical lesions of different grades and lncRNA HOTTIP and H19 single nucleotide polymorphisms[J]. China Oncology, 2022, 32(4): 324-334.
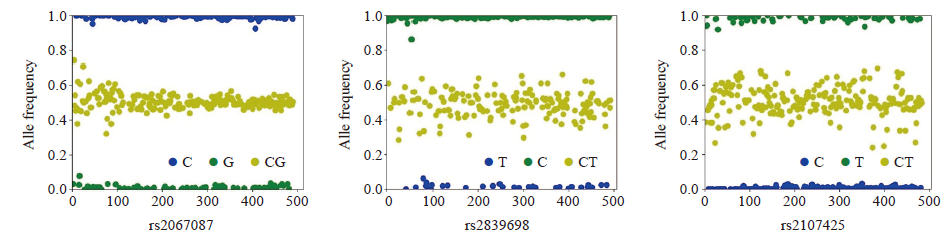
Fig. 1
Genotyping of HOTTIP rs2067087,H19 rs2839698 and H19 rs2107425 The x-axis represents samples number; The y-axis represents the proportion of alleles. Samples with allele frequency of 0-0.2 or 0.8-1.0 were determined to be homozygous, and samples between 0.2-0.8 are determined to be heterozygous."
Tab. 1
Hardy-Weinberg genetic balance test of HOTTIP rs2067087, H19 rs2839698, and H19 rs2107425 polymorphism"
| Group | Genotype | Actual frequencies n (%) | Estimated frequencies n (%) | χ2 | P value |
|---|---|---|---|---|---|
| HOTTIP rs2067087 | CC | 133 (36.9) | 136 (37.8) | 0.535 | 0.465 |
| CG | 177 (49.2) | 170 (47.4) | |||
| GG | 50 (13.9) | 53 (14.8) | |||
| H19 rs2839698 | CC | 195 (54.2) | 193 (53.6) | 0.328 | 0.567 |
| CT | 137 (38.1) | 141 (39.2) | |||
| TT | 28 (7.8) | 26 (7.2) | |||
| H19 rs2107425 | CC | 120 (33.3) | 116 (32.1) | 0.892 | 0.345 |
| CT | 168 (46.7) | 177 (49.0) | |||
| TT | 72 (20.0) | 68 (18.9) |
Tab. 2
Distribution of genotype and allele frequency in cervical lesion group and healthy control group"
| Genotype | Controls n (%) | Cases n (%) | P value | OR | 95% CI |
|---|---|---|---|---|---|
| HOTTIP rs2067087 | |||||
| Codominant | |||||
| CC | 133 (36.9) | 110 (31.9) | 1.000 | ||
| CG | 177 (49.2) | 167 (48.4) | 0.433 | 1.141 | 0.821-1.586 |
| GG | 50 (13.9) | 68 (19.7) | 0.028 | 1.644 | 1.055-2.563 |
| Dominant | |||||
| CC | 133 (36.9) | 110 (31.9) | 1.000 | ||
| CG+GG | 227 (63.1) | 235 (68.1) | 0.158 | 1.252 | 0.917-1.709 |
| Recessive | |||||
| CC+ CG | 310 (86.1) | 277 (80.3) | 1.000 | ||
| GG | 50 (13.9) | 68 (19.7) | 0.039 | 1.522 | 1.021-2.269 |
| Allele | |||||
| C | 443 (61.5) | 387 (56.1) | 1.000 | ||
| G | 277 (38.5) | 303 (43.9) | 0.038 | 1.252 | 1.012-1.549 |
| H19 rs2839698 | |||||
| Codominant | |||||
| CC | 195 (54.2) | 179 (51.9) | 1.000 | ||
| CT | 137 (38.1) | 144 (41.7) | 0.391 | 1.145 | 0.840-1.561 |
| TT | 28 (7.8) | 22 (6.4) | 0.608 | 0.856 | 0.473-1.550 |
| Dominant | |||||
| CC | 195 (54.2) | 179 (51.9) | 1.000 | ||
| CT+TT | 165 (45.8) | 166 (48.1) | 0.544 | 1.096 | 0.815-1.473 |
| Recessive | |||||
| CC+CT | 332 (92.2) | 323 (93.6) | 1.000 | ||
| TT | 28 (7.8) | 22 (6.4) | 0.470 | 0.808 | 0.453-1.441 |
| Allele | |||||
| C | 527 (73.2) | 502 (72.8) | 1.000 | ||
| T | 193 (26.8) | 188 (27.2) | 0.852 | 1.023 | 0.808-1.294 |
| H19 rs2107425 | |||||
| Codominant | |||||
| CC | 120 (33.3) | 114 (33.0) | 1.000 | ||
| CT | 168 (46.7) | 173 (50.1) | 0.635 | 1.084 | 0.777-1.512 |
| TT | 72 (20.0) | 58 (16.8) | 0.453 | 0.848 | 0.551-1.304 |
| Dominant | |||||
| CC | 120 (33.3) | 114 (33.0) | 1.000 | ||
| CT+TT | 240 (66.7) | 231 (67.0) | 0.935 | 1.013 | 0.740-1.386 |
| Recessive | |||||
| CC+CT | 288 (80.0) | 287 (83.2) | 1.000 | ||
| TT | 72 (20.0) | 58 (16.8) | 0.276 | 0.808 | 0.551-1.185 |
| Allele | |||||
| C | 408 (56.7) | 401 (58.1) | 1.000 | ||
| T | 312 (43.3) | 289 (41.9) | 0.582 | 0.942 | 0.763-1.164 |
Tab. 3
Correlation between HOTTIP rs2067087, H19 rs2839698, rs2107425 and cervical lesions at different ages"
| Age/year | Genotype | Controls n (%) | Cases n (%) | P value | OR | 95 % CI | |
|---|---|---|---|---|---|---|---|
| HOTTIP rs2067087 | |||||||
| ≤50 | Dominant | ||||||
| CG+GG | 108 (64.7) | 123 (66.5) | 1.000 | ||||
| CC | 59 (35.3) | 62 (33.5) | 0.720 | 0.923 | 0.594-1.433 | ||
| Recessive | |||||||
| CC+CG | 139 (83.2) | 145 (78.4) | 1.000 | ||||
| GG | 28 (16.8) | 40 (21.6) | 0.250 | 1.369 | 0.801-2.341 | ||
| >50 | Dominant | ||||||
| CG+GG | 119 (61.7) | 112 (70.0) | 1.000 | ||||
| CC | 74 (38.3) | 48 (30.0) | 0.102 | 0.689 | 0.441-1.076 | ||
| Recessive | |||||||
| CC+CG | 171 (88.6) | 132 (82.5) | 1.000 | ||||
| GG | 22 (11.4) | 28 (17.5) | 0.104 | 1.649 | 0.902-3.013 | ||
| H19 rs2839698 | |||||||
| ≤50 | Dominant | ||||||
| CC | 94 (56.3) | 96 (51.9) | 1.000 | ||||
| CT+TT | 73 (43.7) | 89 (48.1) | 0.409 | 1.194 | 0.784-1.817 | ||
| Recessive | |||||||
| CC+CT | 157 (94.0) | 171 (92.4) | 1.000 | ||||
| TT | 10 (6.0) | 14 (7.6) | 0.558 | 1.285 | 0.555-2.977 | ||
| >50 | Dominant | ||||||
| CC | 101 (52.3) | 83 (51.9) | 1.000 | ||||
| CT+TT | 92 (47.7) | 77 (48.1) | 0.932 | 1.018 | 0.670-1.549 | ||
| Recessive | |||||||
| CC+CT | 175 (90.7) | 152 (95.0) | 1.000 | ||||
| TT | 18 (9.3) | 8 (5.0) | 0.127 | 0.512 | 0.216-1.210 | ||
| H19 rs2107425 | |||||||
| ≤50 | Dominant | ||||||
| CC | 49 (29.3) | 64 (34.6) | 1.000 | ||||
| CT+TT | 118 (70.7) | 121 (65.4) | 0.292 | 0.785 | 0.500-1.232 | ||
| Recessive | |||||||
| CC+CT | 127 (76.0) | 153 (82.7) | 1.000 | ||||
| TT | 40 (24.0) | 32 (17.3) | 0.124 | 0.664 | 0.394-1.118 | ||
| >50 | Dominant | ||||||
| CC | 71 (36.8) | 50 (31.3) | 1.000 | ||||
| CT+TT | 122 (63.2) | 110 (68.8) | 0.276 | 1.280 | 0.821-1.996 | ||
| Recessive | |||||||
| CC+CT | 161 (83.4) | 134 (83.8) | 1.000 | ||||
| TT | 32 (16.6) | 26 (16.3) | 0.934 | 0.976 | 0.554-1.719 | ||
Tab. 4
Distribution of genotype and allele frequency of HOTTIP rs2067087, H19 rs2839698, rs2107425 among CIN group, cervical cancer group and control group"
| Genotype | Controls n (%) | CIN n (%) | P value | OR (95% CI) | CC, n % | P value | OR (95% CI) |
|---|---|---|---|---|---|---|---|
| HOTTIP rs2067087 | |||||||
| Codominant | |||||||
| CC | 133 (36.9) | 75 (34.1) | 1.000 | 35 (28.0) | 1.000 | ||
| CG | 177 (49.2) | 97 (44.1) | 0.882 | 0.972 (0.667-1.415) | 70 (56.0) | 0.085 | 1.503 (0.945-2.390) |
| GG | 50 (13.9) | 48 (21.8) | 0.032 | 1.702 (1.046-2.770) | 20 (16.0) | 0.199 | 1.520 (0.803-2.878) |
| Dominant | |||||||
| CC | 133 (36.9) | 75 (34.1) | 1.000 | 35 (28.0) | 1.000 | ||
| CG+GG | 227 (63.1) | 145 (65.9) | 0.487 | 1.133 (0.797-1.610) | 90 (72.0) | 0.071 | 1.507 (0.965-2.352) |
| Recessive | |||||||
| CC+CG | 310 (86.1) | 172 (78.2) | 1.000 | 105 (84.0) | 1.000 | ||
| GG | 50 (13.9) | 48 (21.8) | 0.014 | 1.730 (1.117-2.680) | 20 (16.0) | 0.563 | 1.181 (0.672-2.075) |
| Allele | |||||||
| C | 443 (61.5) | 247 (56.1) | 1.000 | 140 (56.5) | 1.000 | ||
| G | 277 (38.5) | 193 (43.9) | 0.070 | 1.250 (0.982-1.590) | 108 (43.5) | 0.159 | 1.234 (0.921-1.653) |
| H19 rs2839698 | |||||||
| Codominant | |||||||
| CC | 195 (54.2) | 117 (53.2) | 1.000 | 62 (49.6) | 1.000 | ||
| CT | 137 (38.1) | 84 (38.2) | 0.905 | 1.022 (0.686-1.396) | 60 (48.0) | 0.131 | 1.377 (0.908-2.090) |
| TT | 28 (7.8) | 19 (8.6) | 0.700 | 1.131 (0.605-2.115) | 3 (2.4) | 0.082 | 0.337 (0.099-1.147) |
| Dominant | |||||||
| CC | 195 (54.2) | 117 (53.2) | 1.000 | 62 (49.6) | 1.000 | ||
| CT+TT | 165 (45.8) | 103 (46.8) | 0.817 | 1.040 (0.743-1.456) | 63 (50.4) | 0.378 | 1.201 (0.799-1.805) |
| Recessive | |||||||
| CC+CT | 332 (92.2) | 201 (91.4) | 1.000 | 122 (97.6) | 1.000 | ||
| TT | 28 (7.8) | 19 (8.6) | 0.713 | 1.121 (0.610-2.059) | 3 (2.4) | 0.046 | 0.292 (0.087-0.976) |
| Allele | |||||||
| C | 527 (73.2) | 318 (72.3) | 1.000 | 184 (73.6) | 1.000 | ||
| T | 193 (26.8) | 122 (27.7) | 0.732 | 1.048 (0.803-1.367) | 66 (26.4) | 0.901 | 0.979 (0.707-1.357) |
| H19 rs2107425 | |||||||
| Codominant | |||||||
| CC | 120 (33.3) | 66 (30.0) | 1.000 | 48 (38.4) | 1.000 | ||
| CT | 168 (46.7) | 117 (53.2) | 0.226 | 1.266 (0.864-1.855) | 56 (44.8) | 0.428 | 0.833 (0.531-1.308) |
| TT | 72 (20.0) | 37 (16.8) | 0.789 | 0.934 (0.568-1.536) | 21 (16.8) | 0.293 | 0.729 (0.404-1.316) |
| Dominant | |||||||
| CC | 120 (33.3) | 66 (30.0) | 1.000 | 48 (38.4) | 1.000 | ||
| CT+TT | 240 (66.7) | 154 (70.0) | 0.404 | 1.167 (0.812-1.676) | 77 (61.6) | 0.305 | 0.802 (0.526-1.223) |
| Recessive | |||||||
| CC+CT | 288 (80.0) | 183 (83.2) | 1.000 | 104 (83.2) | 1.000 | ||
| TT | 72 (20.0) | 37 (16.8) | 0.342 | 0.809 (0.522-1.253) | 21 (16.8) | 0.434 | 0.808 (0.473-1.380) |
| Allele | |||||||
| C | 408 (56.7) | 249 (56.6) | 1.000 | 152 (60.8) | 1.000 | ||
| T | 312 (43.3) | 191 (43.4) | 0.980 | 1.003 (0.790-1.274) | 98 (39.2) | 0.255 | 0.843 (0.629-1.131) |
Tab. 5
Correlation between HOTTIP rs2067087, H19 rs2839698, rs2107425 and different pathological types of cervical cancer"
| Genotype | Cervical adenocarcinoma n (%) | The squamous cell carcinoma of cervix n (%) | χ2 | P value | OR | 95 % CI |
|---|---|---|---|---|---|---|
| HOTTIP rs2067087 | 0.007 | 0.933 | 0.341-2.685 | |||
| CC | 6 (27.3) | 29 (28.2) | 1.000 | |||
| CG+GG | 16 (72.2) | 74 (71.8) | 0.957 | |||
| H19 rs2839698 | 0.261 | 0.610 | 0.505-3.204 | |||
| CC | 12 (54.5) | 50 (48.5) | 1.000 | |||
| CT+TT | 10 (45.5) | 53 (51.5) | 1.272 | |||
| H19 rs2107425 | 0.047 | 0.829 | 0.346-2.338 | |||
| CC | 8 (36.4) | 40 (38.8) | 1.000 | |||
| CT+TT | 14 (63.6) | 63 (61.2) | 0.900 |
Tab. 6
Correlation between HOTTIP rs2067087, H19 rs2839698, H19 rs2107425 and cervical cancer of different clinical stages"
| Genotype | Phase I n (%) | Phase Ⅱ n (%) | χ2 | P value | OR | 95 % CI | ||
|---|---|---|---|---|---|---|---|---|
| HOTTIP | ||||||||
| rs2067087 | ||||||||
| CC | 9 | 22.0 | 26 | 31.0 | 1.000 | |||
| CG+GG | 32 | 78.0 | 58 | 69.0 | 1.079 | 0.295 | 0.627 | 0.262-1.501 |
| H19 rs2839698 | ||||||||
| CC | 19 | 46.3 | 43 | 51.2 | 1.000 | |||
| CT+TT | 22 | 53.7 | 41 | 48.8 | 0.259 | 0.611 | 0.823 | 0.390-1.740 |
| H19 rs2107425 | ||||||||
| CC | 18 | 43.9 | 30 | 35.7 | 1.000 | |||
| CT+TT | 23 | 56.1 | 54 | 64.3 | 0.778 | 0.378 | 1.409 | 0.658-3.016 |
| [1] |
SUNG H,, FERLAY J,, SIEGEL R L, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71(3): 209-249.
doi: 10.3322/caac.21660 |
| [2] |
XU R R,, ZHANG X Y,, XU Y, et al. Long noncoding RNA MST1P2 promotes cervical cancer progression by sponging with microRNA miR-133b[J]. Bioengineered, 2021, 12(1): 1851-1860.
doi: 10.1080/21655979.2021.1921550 |
| [3] |
KUMARI S,, BHOR V M. Association of cervicovaginal dysbiosis mediated HPV infection with cervical intraepithelial neoplasia[J]. Microb Pathog, 2021, 152: 104780.
doi: 10.1016/j.micpath.2021.104780 |
| [4] |
COHEN P A,, JHINGRAN A,, OAKNIN A, et al. Cervical cancer[J]. Lancet, 2019, 393(10167): 169-182.
doi: 10.1016/S0140-6736(18)32470-X |
| [5] | ELSAYED E T,, SALEM P E,, DARWISH A M, et al. Plasma long non-coding RNA HOTAIR as a potential biomarker for gastric cancer[J]. Int J Biol Markers, 2018: 1724600818760244. |
| [6] | SHENG J Q,, WANG M R,, FANG D, et al. LncRNA NBR2 inhibits tumorigenesis by regulating autophagy in hepatocellular carcinoma[J]. Biomedecine Pharmacother, 2021, 133: 111023. |
| [7] |
CHEN P Y,, HSIEH P L,, PENG C Y, et al. LncRNA MEG3 inhibits self-renewal and invasion abilities of oral cancer stem cells by sponging miR-421[J]. J Formos Med Assoc, 2021, 120(4): 1137-1142.
doi: 10.1016/j.jfma.2020.09.006 |
| [8] |
WANG K C,, YANG Y W,, LIU B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression[J]. Nature, 2011, 472(7341): 120-124.
doi: 10.1038/nature09819 |
| [9] |
RATAJCZAK M Z. Igf2-H19, an imprinted tandem gene, is an important regulator of embryonic development, a guardian of proliferation of adult pluripotent stem cells, a regulator of longevity, and a ‘passkey’ to cancerogenesis[J]. Folia Histochem Cytobiol, 2012, 50(2): 171-179.
doi: 10.5603/FHC.2012.0026 |
| [10] |
OU L,, WANG D Z,, ZHANG H, et al. Decreased expression of miR-138-5p by lncRNA H19 in cervical cancer promotes tumor proliferation[J]. Oncol Res, 2018, 26(3): 401-410.
doi: 10.3727/096504017X15017209042610 |
| [11] | 刘芳. 长链非编码RNA HOTTIP影响宫颈癌细胞增殖、迁移及侵袭能力的研究[D]. 合肥: 安徽大学, 2015. |
| [12] |
WOJCIK G L,, KAO W H L,, DUGGAL P. Relative performance of gene- and pathway-level methods as secondary analyses for genome-wide association studies[J]. BMC Genet, 2015, 16: 34.
doi: 10.1186/s12863-015-0191-2 |
| [13] |
WANG Y C,, TSAO S M,, LI Y T, et al. The relationship between long noncoding RNA H19 polymorphism and the epidermal growth factor receptor phenotypes on the clinicopathological characteristics of lung adenocarcinoma[J]. Int J Environ Res Public Health, 2021, 18(6): 2862.
doi: 10.3390/ijerph18062862 |
| [14] | WANG B G,, LI Y Z,, DING H X, et al. HOTTIP polymorphism may affect gastric cancer susceptibility by altering HOTTIP expression[J]. Biosci Rep, 2020, 40(8): BSR20191687. |
| [15] |
TONG Y S,, WANG X W,, ZHOU X L, et al. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma[J]. Mol Cancer, 2015, 14: 3.
doi: 10.1186/1476-4598-14-3 |
| [16] | YANG J,, WANG Y Y,, ZHANG S, et al. The association of TNF-α promoter polymorphisms with genetic susceptibility to cervical cancer in a Chinese Han population[J]. Int J Gen Med, 2022, 15: 417-427. |
| [17] |
ALI M A,, SHAKER O G,, EZZAT E M, et al. Association between rs1859168/HOTTIP expression level and colorectal cancer and adenomatous polyposis risk in Egyptians[J]. J Interferon Cytokine Res, 2020, 40(6): 279-291.
doi: 10.1089/jir.2019.0105 |
| [18] |
WANG B G,, XU Q,, LV Z, et al. Association of twelve polymorphisms in three onco-lncRNA genes with hepatocellular cancer risk and prognosis: a case-control study[J]. World J Gastroenterol, 2018, 24(23): 2482-2490.
doi: 10.3748/wjg.v24.i23.2482 |
| [19] | LV Z,, XU Q,, SUN L P, et al. Four novel polymorphisms in long non-coding RNA HOTTIP are associated with the risk and prognosis of colorectal cancer[J]. Biosci Rep, 2019, 39(5): BSR20180573. |
| [20] | 于洁,, 卢博奇,, 朱丽业, 等. 长链非编码RNA H19靶向Bcl-2的表达调控宫颈癌细胞侵袭转移行为的机制[J]. 解放军医药杂志, 2019, 31(1): 12-17. |
| YU J,, LU B Q,, ZHU L Y, et al. Mechanism of long-chain non-coding RNA H19 targeting bcl-2 expression for regulating invasion and metastasis of cervical cancer cells[J]. Med Pharm J Chin People’s Liberation Army, 2019, 31(1): 12-17. | |
| [21] |
YANG C,, TANG R,, MA X, et al. Tag SNPs in long non-coding RNA H19 contribute to susceptibility to gastric cancer in the Chinese Han population[J]. Oncotarget, 2015, 6(17): 15311-15320.
doi: 10.18632/oncotarget.3840 |
| [22] | WU E R,, CHOU Y E,, LIU Y F, et al. Association of lncRNA H19 gene polymorphisms with the occurrence of hepatocellular carcinoma[J]. Genes (Basel), 2019, 10(7): E506. |
| [23] |
LIU C H,, CHEN L S,, YOU Z H, et al. Association between lncRNA H19 polymorphisms and cancer susceptibility based on a meta-analysis from 25 studies[J]. Gene, 2020, 729: 144317.
doi: 10.1016/j.gene.2019.144317 |
| [24] | YU B Q,, CHEN J Y,, HOU C F,, et al. LncRNA H 19 gene rs2839698 polymorphism is associated with a decreased risk of colorectal cancer in a Chinese Han population: a case-control study[J]. J Clin Lab Anal, 2020, 34(8): e23311. |
| [25] |
HASHEMI M,, MOAZENI-ROODI A,, SARABANDI S, et al. Association between genetic polymorphisms of long noncoding RNA H19 and cancer risk: a meta-analysis[J]. J Genet, 2019, 98: 81.
doi: 10.1007/s12041-019-1126-x |
| [26] |
HUANG M C,, CHOU Y H,, SHEN H P, et al. The clinicopathological characteristic associations of long non-coding RNA gene H19 polymorphisms with uterine cervical cancer[J]. J Cancer, 2019, 10(25): 6191-6198.
doi: 10.7150/jca.36707 |
| [27] | YANG P J,, HSIEH M J,, HUNG T W, et al. Effects of long noncoding RNA H19 polymorphisms on urothelial cell carcinoma development[J]. Int J Environ Res Public Health, 2019, 16(8): E1322. |
| [28] |
HUANG M C,, CHOU Y H,, SHEN H P, et al. The clinicopathological characteristic associations of long non-coding RNA gene H19 polymorphisms with uterine cervical cancer[J]. J Cancer, 2019, 10(25): 6191-6198.
doi: 10.7150/jca.36707 |
| [1] | CHEN Xun, ZHENG Zhenxia, RUAN Xueru. Effects of TMCO1 on proliferation and migration of cervical cancer cells [J]. China Oncology, 2024, 34(6): 571-580. |
| [2] | FENG Zheng, GUO Qinhao, ZHU Jun, WU Xiaohua, WEN Hao. Progress in treatment of gynecological cancer in 2023 [J]. China Oncology, 2024, 34(4): 340-360. |
| [3] | SHEN Jie, FENG Xiaoshuang, WEN Hao, ZHOU Changming, MO Miao, WANG Zezhou, YUAN Jing, WU Xiaohua, ZHENG Ying. Metastasis patterns and survival analysis of 572 patients with metastatic cervical cancer: a hospital-based real world study [J]. China Oncology, 2024, 34(4): 361-367. |
| [4] | XIA Lingfang, ZHU Jun, WU Xiaohua. The latest progress and prospect of gynecological tumor treatment at 2023 ESMO [J]. China Oncology, 2023, 33(11): 969-980. |
| [5] | GUO Qinhao, YU Min, WU Xiaohua. Progress in diagnosis and treatment of gynecological tumors in 2022 [J]. China Oncology, 2023, 33(1): 14-24. |
| [6] | CHEN Anli, SHEN Haoyuan, YANG Shixiong, DENG Chunyan, HU Chaohua, LIU Hanzhong, WANG Shu, QIAN Fang. Predictive value of plasma long non-coding RNA H19 in neoadjuvant therapy for breast cancer [J]. China Oncology, 2022, 32(8): 727-735. |
| [7] | PANG Yi, WU Chunxiao, GU Kai, BAO Pingping, WANG Chunfang, SHI Liang, GONG Yangming, XIANG Yongmei, DOU Jianming, WU Mengyin, FU Chen, SHI Yan. Analysis of current status of cervical cancer incidence and mortality in Shanghai, 2016 and trends of 2002-2016 [J]. China Oncology, 2022, 32(6): 519-526. |
| [8] | FANG Yanhui , MA Caijuan , ZHANG Chunli , ZHAI Jiawei , ZHU Qiaoying , LIU Yin , ZHAO Xiwa . miR-496 inhibits proliferation, migration and invasion of HeLa cell in cervical cancer via SFMBT1 [J]. China Oncology, 2021, 31(8): 697-703. |
| [9] | LONG Xingtao , ZHOU Qi , WANG Dong , CHEN Yuemei , JIN Fujun . The prognostic value of revised 2018 FIGO stage ⅢC in cervical cancer [J]. China Oncology, 2021, 31(8): 725-733. |
| [10] | YANG Jia , LIU Shuyuan , WANG Yingying , LI Yingfu , MA Qianli , LI Yu , WANG Yongrong , LI Chuanyin , TAN Fang . The association of polymorphisms in TNF-α gene with non-small cell lung cancer in Han population of Yunnan Province [J]. China Oncology, 2021, 31(7): 616-628. |
| [11] | DONG Shijie, HU Xiaoxin, WANG Wei, YANG Meng, YUE Lei, TONG Tong, GU Yajia. Prediction of lymph node metastasis of cervical cancer based on multi-sequence MRI and multi-system imaging omics model [J]. China Oncology, 2021, 31(6): 460-467. |
| [12] | ZHU Jun, WU Xiaohua. Leading research progress and prospect of gynecological oncology in 2020 [J]. China Oncology, 2021, 31(4): 250-256. |
| [13] | XU Shanshan , SONG Qinqin , WU Hongjiao , FU Ning , CHE Lingyu , JIN Min , LIU Chong , HAN Sugui , ZHANG Xuemei , ZHANG Zhi . Genetic variation in membrane cofactor protein CD46 rs1970530 affects the risk of hepatocellular carcinoma [J]. China Oncology, 2020, 30(9): 656-660. |
| [14] | SHEN Jie , MO Miao , YUAN Jing , ZHOU Changming , WANG Zezhou , ZHANG Zhihong , WEN Hao , WU Xiaohua , ZHENG Ying . Long-term survival report on 1 313 patients with locally advanced cervical cancer: a hospital-based real world study [J]. China Oncology, 2020, 30(3): 192-198. |
| [15] | CHEN Xiaohui, WANG Jiazhou, HU Weigang, PENG Jiayuan, ZHAI Peng. Dosimetric impact of machine and treatment planning system on knowledge-based planning: a study based on cervical cancer IMRT plan [J]. China Oncology, 2020, 30(10): 821-825. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
沪ICP备12009617
Powered by Beijing Magtech Co. Ltd
