Welcome to China Oncology,
China Oncology ›› 2023, Vol. 33 ›› Issue (5): 484-498.doi: 10.19401/j.cnki.1007-3639.2023.05.009
• Article • Previous Articles Next Articles
YANG Wenxiao1,2( ), GUO Linwei3, LING Hong1, HU Xin1,2(
), GUO Linwei3, LING Hong1, HU Xin1,2( )
)
Received:2023-01-31
Revised:2023-04-26
Online:2023-05-30
Published:2023-06-16
Contact:
HU Xin
Share article
CLC Number:
YANG Wenxiao, GUO Linwei, LING Hong, HU Xin. Characterization of immune microenvironment identifies prognostic and immunotherapy benefit for trastuzumab-based therapy[J]. China Oncology, 2023, 33(5): 484-498.

Fig. 3
Immune deficiency in trastuzumab-resistant patients A, B: Volcano plot of RNA-seq gene expression changes in the trastuzumab-resistant neoadjuvant and adjuvant cohorts. C, D: KEGG pathway analysis in the trastuzumab-resistant neoadjuvant and adjuvant cohorts. E, F: Top 10 candidate GO terms and pathways in the functional annotation of GSEA of the trastuzumab-resistant neoadjuvant and adjuvant cohorts. G: Representative pathways of GSEA in the trastuzumab-resistant group (P-adjust<0.05) of neoadjuvant and adjuvant cohorts.."
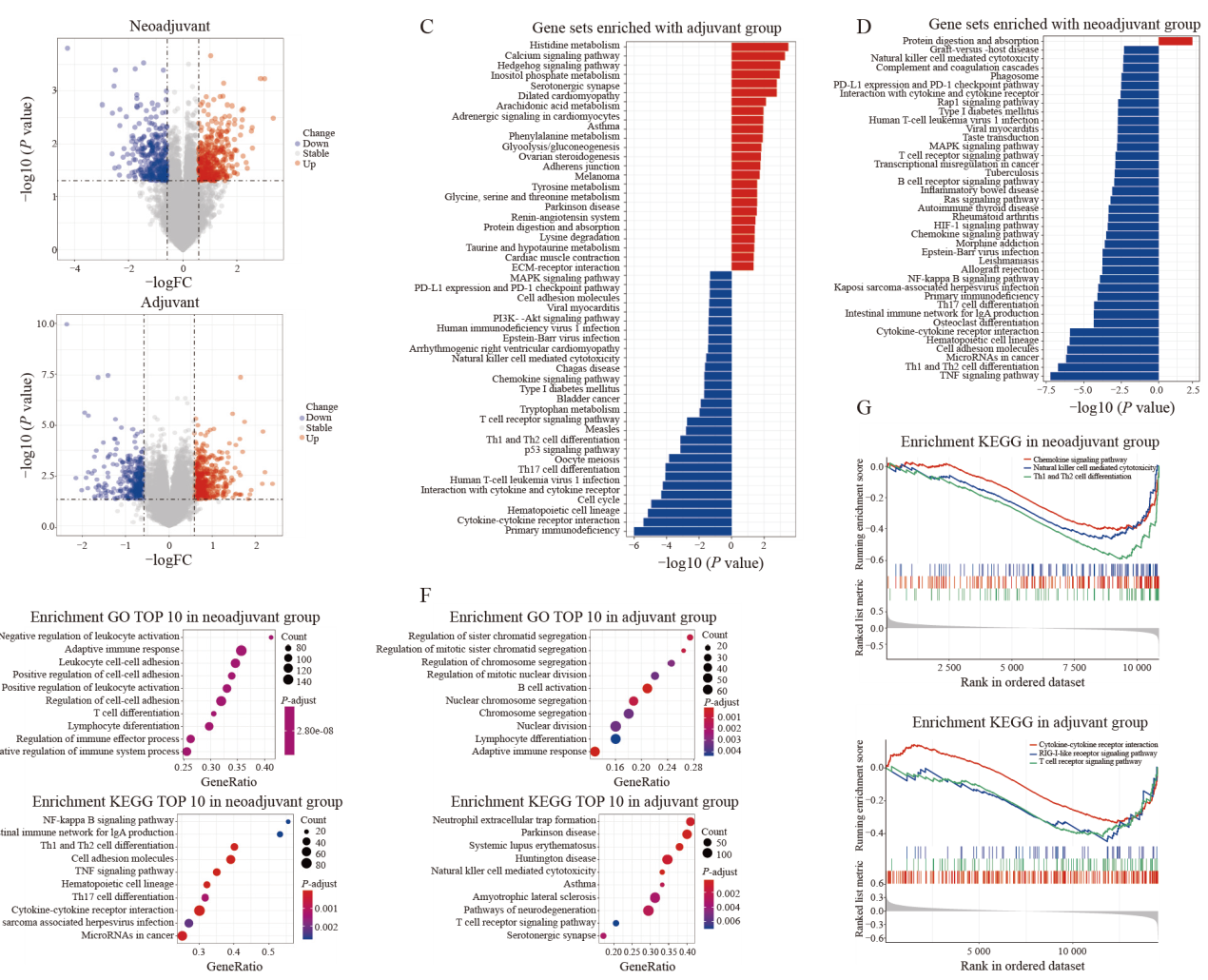
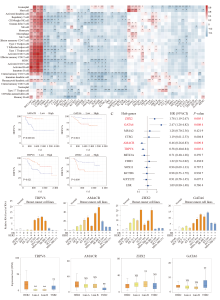
Fig. 5
Trastuzumab-response related hub genes A: Correlation coefficient heatmap to demonstrate the immune characteristics of the hub genes. B: Kaplan-Meier analysis of the DDFS curves for the four genes (of the 12 hub genes) with significant prognostic value (log-rank test, P<0.05). C: Univariate COX regression analysis of the 12 hub genes. D: RTFQ-PCR validation of the four prognostic genes in breast cancer cell lines. E. Expression analysis of the four prognostic genes in TCGA cohort. Significant differences between the two subgroups were assessed using the Wilcoxon test (NS: Not significant; *: P<0.05; **: P<0.01)."
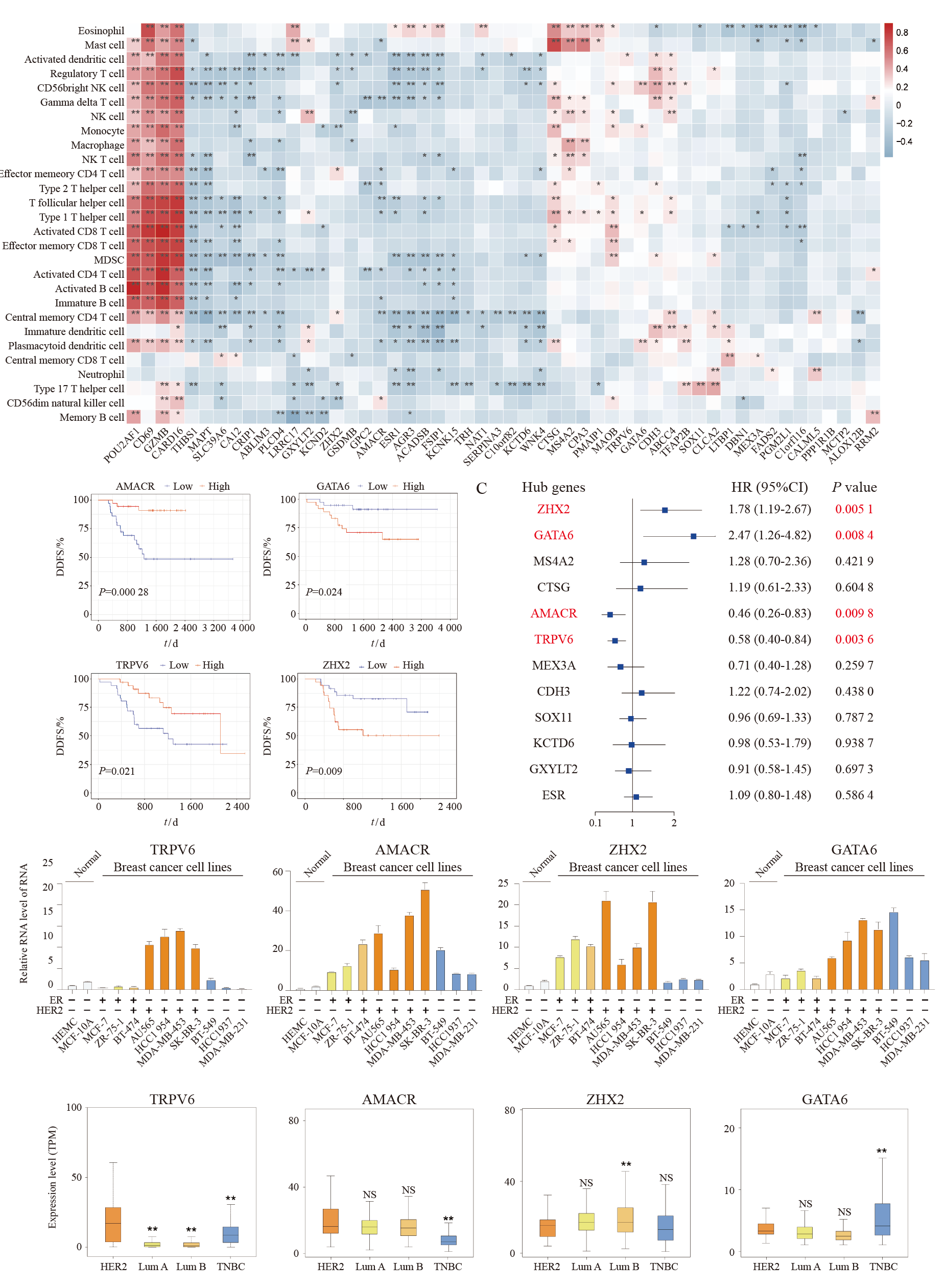
Fig. 6
TRGPI construction and evaluation Multivariate COX regression analysis of clinicopathological factors and TRGPI in the training set (A) and test set (B). C: Kaplan-Meier survival analysis of the TRGPI subgroups in the training set (left) and test set (right) (log-rank test, P<0.05). D: ROC curve analysis of the prognostic value of TRGPI for DDFS at 18 months and 36 months in the training set (left) and the test set (right). E: The TRGPI score curve showing the distribution of patients under trastuzumab therapy in the test set. Significant differences between the two subgroups were assessed using the Wilcoxon test (*: P<0.05; ***: P<0.001)."
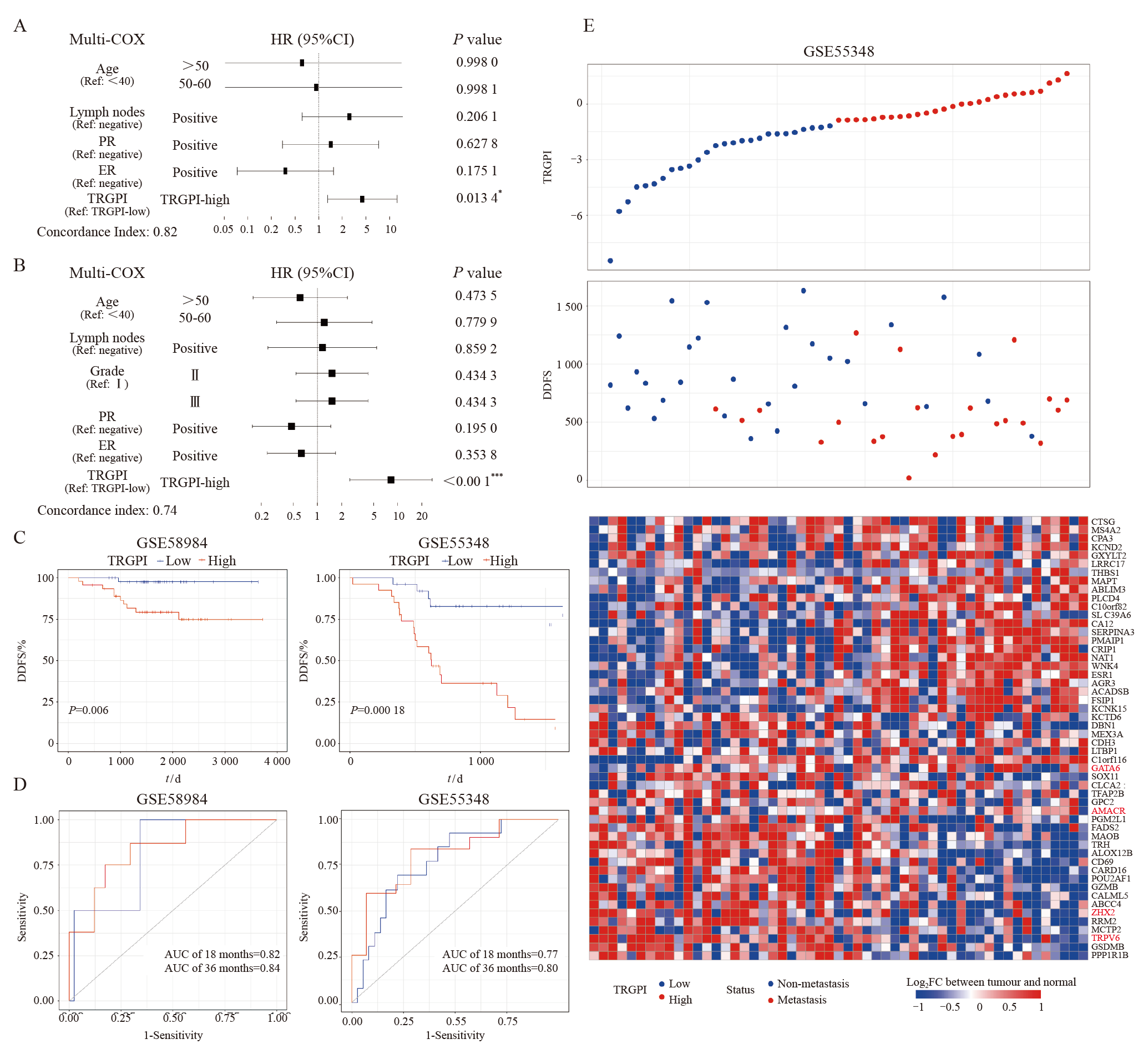
Fig. 7
Genomic features in TRGPI subgroups A: Waterfall plots of 30 highly variant mutant genes demonstrated the mutation landscape in the TRGPI-low group (left) and TRGPI-high group (right). B: Distribution of the top 10 somatic mutations VAF in the TRGPI-low group (upper) and TRGPI-high group (lower). C: The mutation co-occurrence and exclusion analyses in TRGPI-low group (upper left) and TRGPI-high group (lower right). NS: No significance. D: Differences in TMB among different TRGPI subgroups. Wilcoxon test was used to compare the statistical difference (NS: Not significant)."
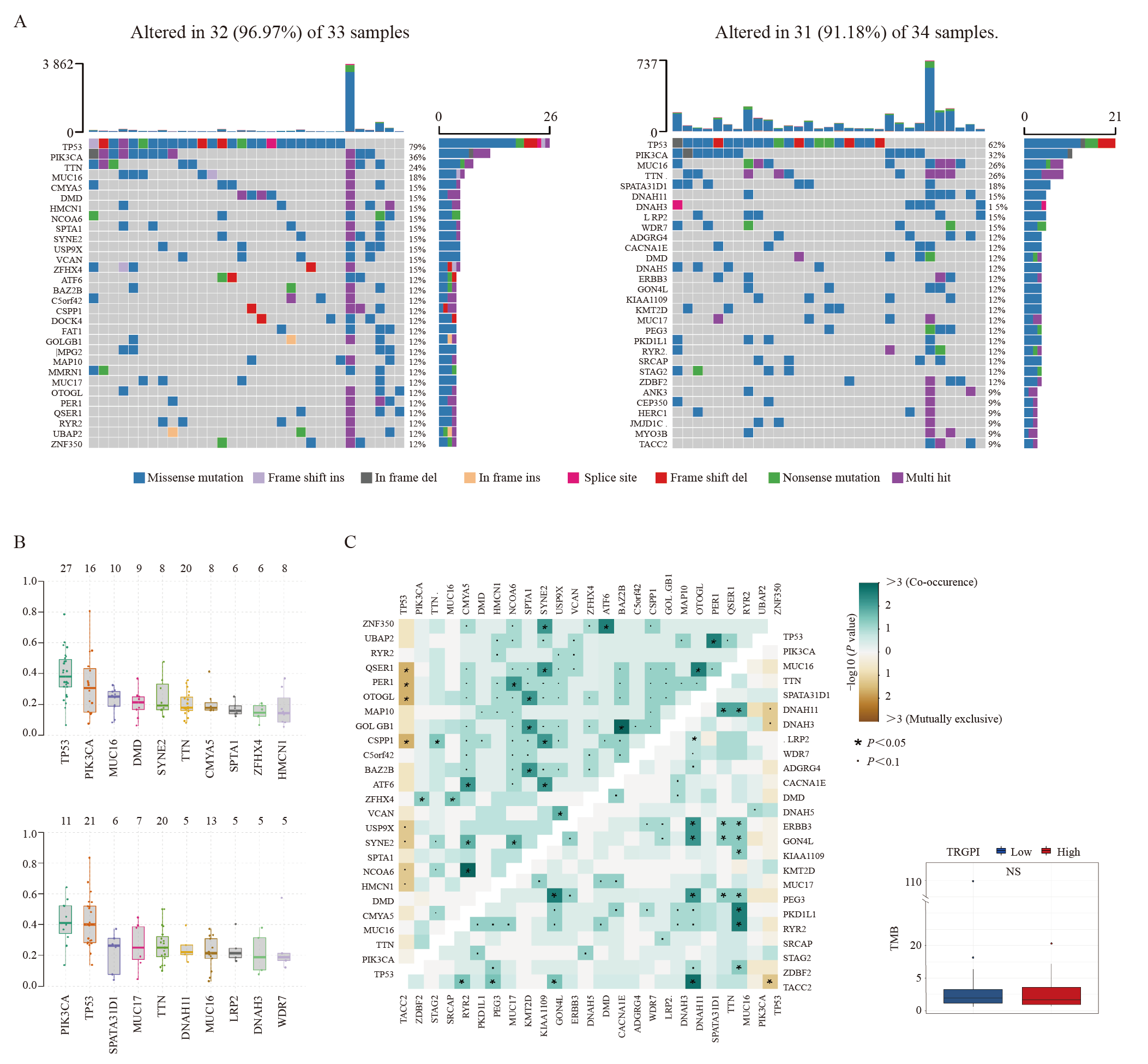
Fig. 8
TIME characteristics in different TRGPI subgroups of the training set A: The proportions of TIME cells in different TRGPI subgroups. Significant differences between the two subgroups were assessed using the Wilcoxon test. B: The TRGPI grouping and proportions of TIME cells for 91 patients. Age, ER status, PR status, metastasis, DDFS, and TRGPI were used as patient annotations. NS: Not significant; *: P<0.05; ***: P<0.001."
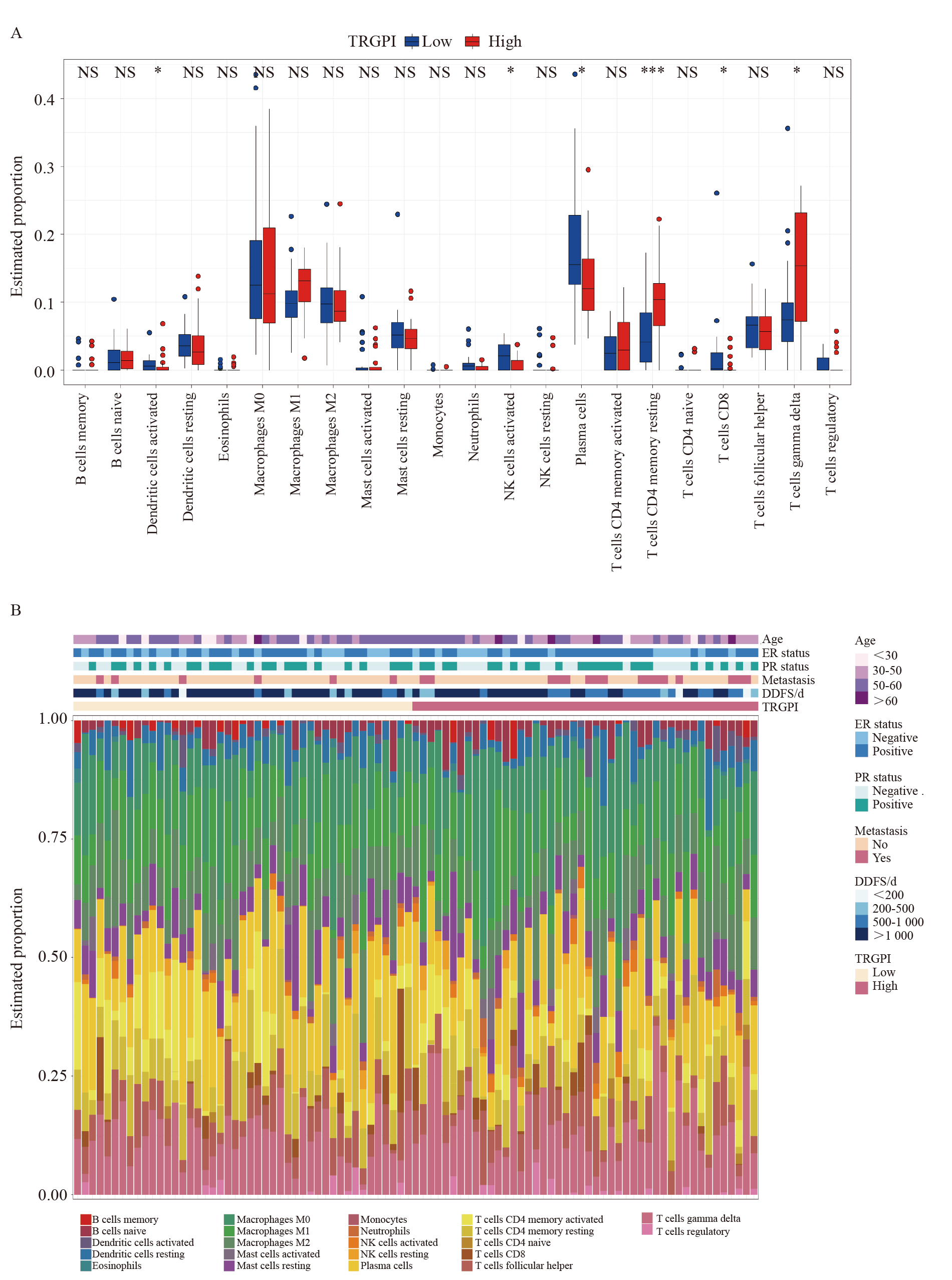
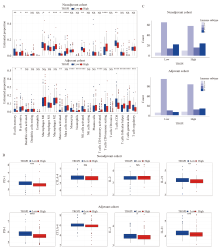
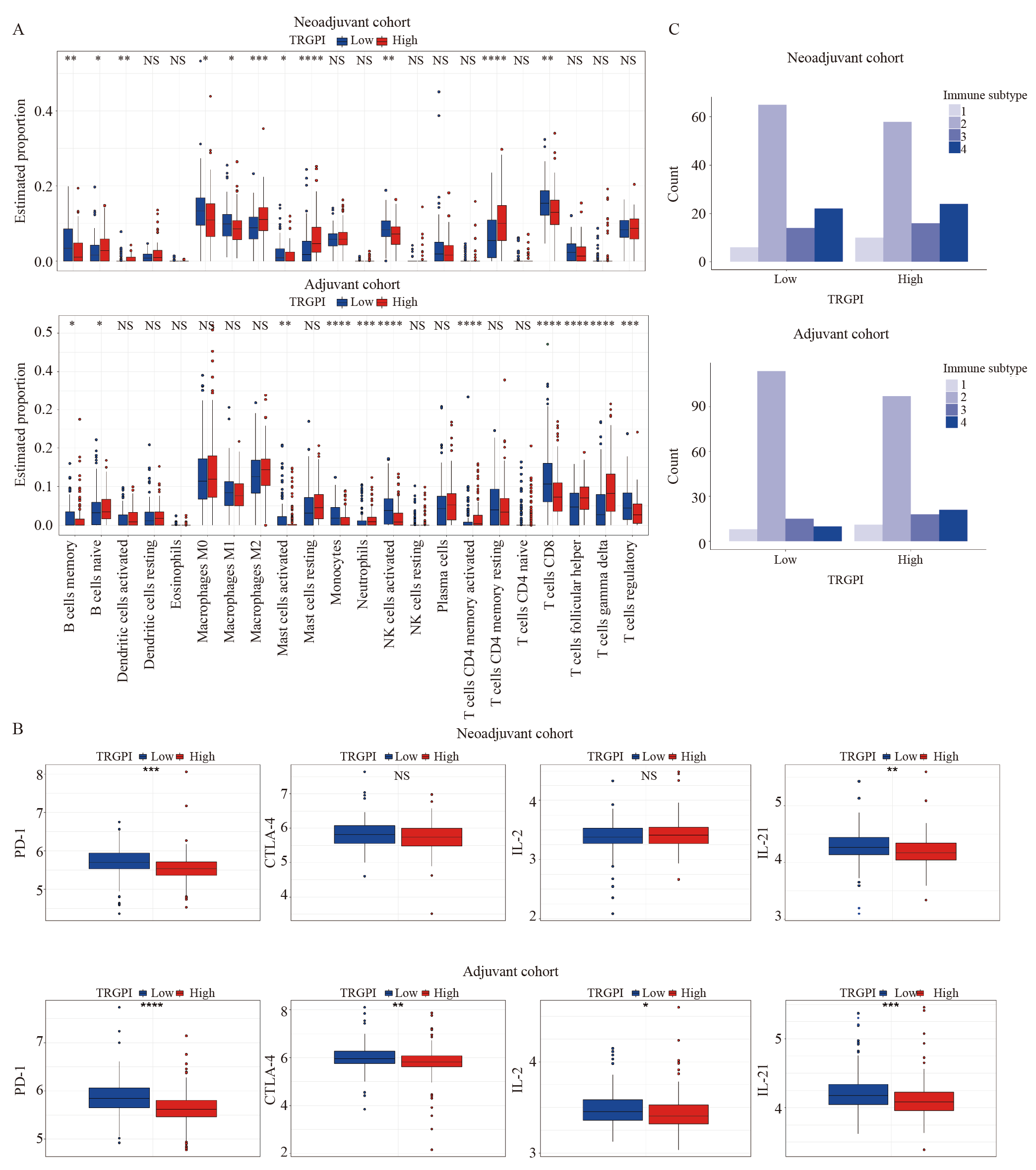
Fig. 9
TIME characteristics of TRGPI subgroups in neoadjuvant and adjuvant cohorts A: The percentage of immune cells; B:The expression levels of PD-1, CTLA-4, IL-2, and IL-21 from different TRGPI subgroups were compared in the neoadjuvant and adjuvant cohorts. Significant differences between the two subgroups were assessed using the Wilcoxon test. C: Immune subtype analysis of TRGPI subgroups in the neoadjuvant and adjuvant cohorts. NS: Not significant; *: P<0.05; **: P<0.01; ***: P<0.001; ****:P<0.000 1.."
| [1] |
SLAMON D J, CLARK G M, WONG S G, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene[J]. Science, 1987, 235(4785): 177-182.
doi: 10.1126/science.3798106 pmid: 3798106 |
| [2] |
SWAIN S M, SHASTRY M, HAMILTON E. Targeting HER2-positive breast cancer: advances and future directions[J]. Nat Rev Drug Discov, 2023, 22(2): 101-126.
doi: 10.1038/s41573-022-00579-0 |
| [3] |
SLAMON D J, LEYLAND-JONES B, SHAK S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2[J]. N Engl J Med, 2001, 344(11): 783-792.
doi: 10.1056/NEJM200103153441101 |
| [4] |
VOGEL C L, COBLEIGH M A, TRIPATHY D, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer[J]. J Clin Oncol, 2002, 20(3): 719-726.
doi: 10.1200/JCO.2002.20.3.719 pmid: 11821453 |
| [5] |
GENNARI R, MENARD S, FAGNONI F, et al. Pilot study of the mechanism of action of preoperative trastuzumab in patients with primary operable breast tumors overexpressing HER2[J]. Clin Cancer Res, 2004, 10(17): 5650-5655.
doi: 10.1158/1078-0432.CCR-04-0225 pmid: 15355889 |
| [6] |
BUSSOLATI G, MONTEMURRO F, RIGHI L, et al. A modified trastuzumab antibody for the immunohistochemical detection of HER-2 overexpression in breast cancer[J]. Br J Cancer, 2005, 92(7): 1261-1267.
doi: 10.1038/sj.bjc.6602507 |
| [7] |
DIERMEIER S, HORVÁTH G, KNUECHEL-CLARKE R, et al. Epidermal growth factor receptor coexpression modulates susceptibility to herceptin in HER2/neu overexpressing breast cancer cells via specific erbB-receptor interaction and activation[J]. Exp Cell Res, 2005, 304(2): 604-619.
doi: 10.1016/j.yexcr.2004.12.008 pmid: 15748904 |
| [8] |
LU Y, ZI X, ZHAO Y, et al. Insulin-like growth factor-Ⅰ receptor signaling and resistance to trastuzumab (Herceptin)[J]. J Natl Cancer Inst, 2001, 93(24): 1852-1857.
doi: 10.1093/jnci/93.24.1852 |
| [9] |
NAGATA Y, LAN K H, ZHOU X Y, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients[J]. Cancer Cell, 2004, 6(2): 117-127.
doi: 10.1016/j.ccr.2004.06.022 pmid: 15324695 |
| [10] |
LOI S, GIOBBIE-HURDER A, GOMBOS A, et al. Pembrolizumab plus trastuzumab in trastuzumab-resistant, advanced, HER2-positive breast cancer (PANACEA): a single-arm, multicentre, phase 1b-2 trial[J]. Lancet Oncol, 2019, 20(3): 371-382.
doi: S1470-2045(18)30812-X pmid: 30765258 |
| [11] |
STAGG J, LOI S, DIVISEKERA U, et al. Anti-ErbB-2 MAb therapy requires type Ⅰ and Ⅱ interferons and synergizes with anti-PD-1 or anti-CD137 MAb therapy[J]. Proc Natl Acad Sci U S A, 2011, 108(17): 7142-7147.
doi: 10.1073/pnas.1016569108 |
| [12] |
BASELGA J, BRADBURY I, EIDTMANN H, et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): a randomised, open-label, multicentre, phase 3 trial[J]. Lancet, 2012, 379(9816): 633-640.
doi: 10.1016/S0140-6736(11)61847-3 pmid: 22257673 |
| [13] | CAREY L A, BERRY D A, CIRRINCIONE C T, et al. Molecular heterogeneity and response to neoadjuvant human epidermal growth factor receptor 2 targeting in CALGB 40601, a randomized phase Ⅲ trial of paclitaxel plus trastuzumab with or without lapatinib[J]. J Clin Oncol, 2016, 34(6): 542-549. |
| [14] |
GIANNI L, PIENKOWSKI T, IM Y H, et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): a randomised multicentre, open-label, phase 2 trial[J]. Lancet Oncol, 2012, 13(1): 25-32.
doi: 10.1016/S1470-2045(11)70336-9 pmid: 22153890 |
| [15] |
SCHNEEWEISS A, CHIA S, HICKISH T, et al. Pertuzumab plus trastuzumab in combination with standard neoadjuvant anthracycline-containing and anthracycline-free chemotherapy regimens in patients with HER2-positive early breast cancer: a randomized phase Ⅱ cardiac safety study (TRYPHAENA)[J]. Ann Oncol, 2013, 24(9): 2278-2284.
doi: 10.1093/annonc/mdt182 |
| [16] | GIORDANO S H, FRANZOI M A B, TEMIN S, et al. Systemic therapy for advanced human epidermal growth factor receptor 2-positive breast cancer: ASCO guideline update[J]. J Clin Oncol, 2022, 40(23): 2612-2635. |
| [17] | FU Z W, LI S J, HAN S F, et al. Antibody drug conjugate: the “biological missile” for targeted cancer therapy[J]. Signal Transduct Target Ther, 2022, 7(1): 93. |
| [18] |
TORRES E T R, EMENS L A. Emerging combination immunotherapy strategies for breast cancer: dual immune checkpoint modulation, antibody-drug conjugates and bispecific antibodies[J]. Breast Cancer Res Treat, 2022, 191(2): 291-302.
doi: 10.1007/s10549-021-06423-0 |
| [19] |
FERNANDEZ-MARTINEZ A, PASCUAL T, SINGH B, et al. Prognostic and predictive value of immune-related gene expression signatures vs tumor-infiltrating lymphocytes in early-stage ERBB2/HER2-positive breast cancer: a correlative analysis of the CALGB 40601 and PAMELA trials[J]. JAMA Oncol, 2023, 9(4): 490-499.
doi: 10.1001/jamaoncol.2022.6288 |
| [20] |
SHARMA P, HU-LIESKOVAN S, WARGO J A, et al. Primary, adaptive, and acquired resistance to cancer immunotherapy[J]. Cell, 2017, 168(4): 707-723.
doi: S0092-8674(17)30065-X pmid: 28187290 |
| [21] |
KALAORA S, NAGLER A, WARGO J A, et al. Mechanisms of immune activation and regulation: lessons from melanoma[J]. Nat Rev Cancer, 2022, 22(4): 195-207.
doi: 10.1038/s41568-022-00442-9 pmid: 35105962 |
| [22] |
LABRIE M, BRUGGE J S, MILLS G B, et al. Therapy resistance: opportunities created by adaptive responses to targeted therapies in cancer[J]. Nat Rev Cancer, 2022, 22(6): 323-339.
doi: 10.1038/s41568-022-00454-5 pmid: 35264777 |
| [23] |
COOLEY S, BURNS L J, REPKA T, et al. Natural killer cell cytotoxicity of breast cancer targets is enhanced by two distinct mechanisms of antibody-dependent cellular cytotoxicity against LFA-3 and HER2/neu[J]. Exp Hematol, 1999, 27(10): 1533-1541.
doi: 10.1016/s0301-472x(99)00089-2 pmid: 10517495 |
| [24] |
LEWIS G D, FIGARI I, FENDLY B, et al. Differential responses of human tumor cell lines to anti-p185HER2 monoclonal antibodies[J]. Cancer Immunol Immunother, 1993, 37(4): 255-263.
doi: 10.1007/BF01518520 |
| [25] |
KOHRT H E, HOUOT R, MARABELLE A, et al. Combination strategies to enhance antitumor ADCC[J]. Immunotherapy, 2012, 4(5): 511-527.
doi: 10.2217/imt.12.38 pmid: 22642334 |
| [26] |
CLYNES R A, TOWERS T L, PRESTA L G, et al. Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets[J]. Nat Med, 2000, 6(4): 443-446.
doi: 10.1038/74704 |
| [27] |
SALGADO R, DENKERT C, CAMPBELL C, et al. Tumor-infiltrating lymphocytes and associations with pathological complete response and event-free survival in HER2-positive early-stage breast cancer treated with lapatinib and trastuzumab: a secondary analysis of the NeoALTTO trial[J]. JAMA Oncol, 2015, 1(4): 448-454.
doi: 10.1001/jamaoncol.2015.0830 pmid: 26181252 |
| [28] |
CORTAZAR P, ZHANG L J, UNTCH M, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis[J]. Lancet, 2014, 384(9938): 164-172.
doi: 10.1016/S0140-6736(13)62422-8 pmid: 24529560 |
| [29] |
INGOLD HEPPNER B, UNTCH M, DENKERT C, et al. Tumor-infiltrating lymphocytes: a predictive and prognostic biomarker in neoadjuvant-treated HER2-positive breast cancer[J]. Clin Cancer Res, 2016, 22(23): 5747-5754.
pmid: 27189162 |
| [30] |
CHIC N, LUEN S J, NUCIFORO P, et al. Tumor cellularity and infiltrating lymphocytes as a survival surrogate in HER2-positive breast cancer[J]. J Natl Cancer Inst, 2022, 114(3): 467-470.
doi: 10.1093/jnci/djab057 |
| [31] |
DENKERT C, VON MINCKWITZ G, BRASE J C, et al. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers[J]. J Clin Oncol, 2015, 33(9): 983-991.
doi: 10.1200/JCO.2014.58.1967 pmid: 25534375 |
| [32] | RITCHIE M E, PHIPSON B, WU D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies[J]. Nucleic Acids Res, 2015, 43(7): e47. |
| [33] |
CHALMERS Z R, CONNELLY C F, FABRIZIO D, et al. Analysis of 100 000 human cancer genomes reveals the landscape of tumor mutational burden[J]. Genome Med, 2017, 9(1): 34.
doi: 10.1186/s13073-017-0424-2 |
| [34] |
NEWMAN A M, LIU C L, GREEN M R, et al. Robust enumeration of cell subsets from tissue expression profiles[J]. Nat Methods, 2015, 12(5): 453-457.
doi: 10.1038/nmeth.3337 pmid: 25822800 |
| [35] |
THORSSON V, GIBBS D L, BROWN S D, et al. The immune landscape of cancer[J]. Immunity, 2018, 48(4): 812-830.e14.
doi: S1074-7613(18)30121-3 pmid: 29628290 |
| [36] |
LAUSS M, DONIA M, HARBST K, et al. Mutational and putative neoantigen load predict clinical benefit of adoptive T cell therapy in melanoma[J]. Nat Commun, 2017, 8(1): 1738.
doi: 10.1038/s41467-017-01460-0 pmid: 29170503 |
| [37] |
JIANG P, GU S Q, PAN D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response[J]. Nat Med, 2018, 24(10): 1550-1558.
doi: 10.1038/s41591-018-0136-1 pmid: 30127393 |
| [38] |
SCHOENFELD A J, RIZVI H, BANDLAMUDI C, et al. Clinical and molecular correlates of PD-L1 expression in patients with lung adenocarcinomas[J]. Ann Oncol, 2020, 31(5): 599-608.
doi: S0923-7534(20)36018-X pmid: 32178965 |
| [39] |
JENSEN J D, KNOOP A, LAENKHOLM A V, et al. PIK3CA mutations, PTEN, and pHER2 expression and impact on outcome in HER2-positive early-stage breast cancer patients treated with adjuvant chemotherapy and trastuzumab[J]. Ann Oncol, 2012, 23(8): 2034-2042.
doi: S0923-7534(19)38088-3 pmid: 32018458 |
| [40] |
VIGANO S, ALATZOGLOU D, IRVING M, et al. Targeting adenosine in cancer immunotherapy to enhance T-cell function[J]. Front Immunol, 2019, 10: 925.
doi: 10.3389/fimmu.2019.00925 pmid: 31244820 |
| [41] |
YAMAMOTO K, VENIDA A, YANO J, et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I[J]. Nature, 2020, 581(7806): 100-105.
doi: 10.1038/s41586-020-2229-5 |
| [42] |
PRAT A, GUARNERI V, PASCUAL T, et al. Development and validation of the new HER2DX assay for predicting pathological response and survival outcome in early-stage HER2-positive breast cancer[J]. EBioMedicine, 2022, 75: 103801.
doi: 10.1016/j.ebiom.2021.103801 |
| [43] |
FERNANDEZ-MARTINEZ A, KROP I E, HILLMAN D W, et al. Survival, pathologic response, and genomics in CALGB 40601 (alliance), a neoadjuvant phase Ⅲ trial of paclitaxel-trastuzumab with or without lapatinib in HER2-positive breast cancer[J]. J Clin Oncol, 2020, 38(35): 4184-4193.
doi: 10.1200/JCO.20.01276 |
| [44] |
WONG H, LEUNG R, KWONG A, et al. Integrating molecular mechanisms and clinical evidence in the management of trastuzumab resistant or refractory HER-2+ metastatic breast cancer[J]. Oncologist, 2011, 16(11): 1535-1546.
doi: 10.1634/theoncologist.2011-0165 |
| [45] |
MITTAL D, CARAMIA F, MICHIELS S, et al. Improved treatment of breast cancer with anti-HER2 therapy requires interleukin-21 signaling in CD8+ T cells[J]. Cancer Res, 2016, 76(2): 264-274.
doi: 10.1158/0008-5472.CAN-15-1567 pmid: 26744522 |
| [46] |
KILLOCK D. Targeted therapy: leveraging ADCC to enhance anti-HER2 therapy[J]. Nat Rev Clin Oncol, 2017, 14(4): 200.
doi: 10.1038/nrclinonc.2017.19 pmid: 28195234 |
| [47] |
ROSENBERG S A, YANG J C, SHERRY R M, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy[J]. Clin Cancer Res, 2011, 17(13): 4550-4557.
doi: 10.1158/1078-0432.CCR-11-0116 pmid: 21498393 |
| [48] |
VESELY M D, KERSHAW M H, SCHREIBER R D, et al. Natural innate and adaptive immunity to cancer[J]. Annu Rev Immunol, 2011, 29: 235-271.
doi: 10.1146/annurev-immunol-031210-101324 pmid: 21219185 |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
沪ICP备12009617
Powered by Beijing Magtech Co. Ltd
