Welcome to China Oncology,
China Oncology ›› 2024, Vol. 34 ›› Issue (10): 915-930.doi: 10.19401/j.cnki.1007-3639.2024.10.002
• Article • Previous Articles Next Articles
ZHAO Junxiu1,2( ), ZHU Yi3, SONG Xiaoyu4, ZHE Chao1, XIAO Yuhan1, LIU Yunduo1, LI Linhai2(
), ZHU Yi3, SONG Xiaoyu4, ZHE Chao1, XIAO Yuhan1, LIU Yunduo1, LI Linhai2( ), XIAO Bin2,5(
), XIAO Bin2,5( )
)
Received:2024-03-18
Revised:2024-09-10
Online:2024-10-30
Published:2024-11-20
Contact:
LI Linhai, XIAO Bin
Share article
CLC Number:
ZHAO Junxiu, ZHU Yi, SONG Xiaoyu, ZHE Chao, XIAO Yuhan, LIU Yunduo, LI Linhai, XIAO Bin. Circ-0007766 acts as a miR-1972 sponge to promote breast cancer cell migration and invasion via upregulation of HER2[J]. China Oncology, 2024, 34(10): 915-930.
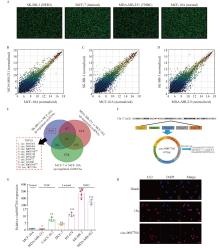
Fig. 1
Circ-0007766 expression increased within HER2-positive breast cancer cells A: The original images of circRNA expression in different breast cancer cells were shown by circRNA microarray. B: Scatter plots showed the differential expression of circRNA in MDA-MB-231 and MCF-10A cells. C: Scatter plots showed the differential expression of circRNA in MCF-7 and MCF-10A cells. D: Scatter plots showed the differential expression of circRNA in SK-BR-3 and MCF-10A cells. E: A Venn diagram showed the differential expression of circRNAs in different breast cancer cells and normal breast cancer cells. F: Chromosomal location and loop formation of circ-0007766. G: The expression of circ-0007766 was determined by RTFQ-PCR in 6 breast cancer cell lines and MCF-10A cells. H: Circ-0007766 was detected by FISH in the subcellular location of SK-BR-3 cells. *: P<0.05, compared with the normal breast cell MCF-10A group; **: P<0.01, compared with the normal breast cell MCF-10A group;***: P<0.001 compared with the normal breast cell MCF-10A group."
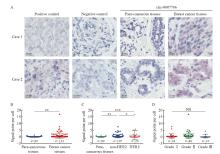
Fig. 2
Circ-0007766 showed high expression within HER2 breast cancer tissues A: BaseScope displayed representative images of the expression and localization of circ-0007766 in breast cancer and para-cancerous tissue. B: Scatter plot of circ-0007766 expression in breast cancer and para-cancerous tissues. C: Scatter plot of circ-0007766 levels in HER2-positive and HER2-negative breast cancer tissues. D: Scatter plot of circ-0007766 expression in breast cancer of different clinical grades. *: P<0.05, compared with non-HER2; **: P<0.01, compared with each other; ***: P<0.001, compared with each other; NS: Without statistically significant meaning."
Tab. 1
BaseScope detection of circ-0007766 expression in tissue"
| Pathological features | Case n | circ-0007766 signal site | P value | Pathological features | Case n | circ-0007766 signal site | P value | |
|---|---|---|---|---|---|---|---|---|
| Types | Tumor diameter D/cm3 | |||||||
| Normal | 89 | 0.051 (0.000, 0.313) | 0.013 | <20 | 84 | 0.185 (0.036, 0.619) | 0.084 | |
| TNBC | 32 | 0.083 (0.006, 0.359) | 0.075 | ≥20 | 51 | 0.101 (0.000, 0.382) | ||
| Luminal A | 47 | 0.106 (0.014, 0.392) | 0.042 | T phase of TNM | ||||
| Luminal B | 28 | 0.288 (0.031, 0.892) | 0.827 | T1 | 29 | 0.168 (0.009, 0.500) | 0.733 | |
| HER2 positive | 26 | 0.253 (0.068, 1.016) | 0.001 | T2 | 91 | 0.169 (0.034, 0.540) | 0.028 | |
| Age/year | T3 | 15 | 0.064 (0.000, 0.228) | 0.104 | ||||
| <54 | 75 | 0.168 (0.029, 0.473) | 0.747 | N phase of TNM | ||||
| ≥54 | 60 | 0.127 (0.027, 0.537) | N0 | 47 | 0.169 (0.029, 0.504) | 0.763 | ||
| Pathological staging | N1 | 41 | 0.143 (0.033, 0.592) | 0.836 | ||||
| Ⅰ | 34 | 0.220 (0.000, 0.510) | 0.724 | N2 | 35 | 0.135 (0.021, 0.383) | 0.675 | |
| Ⅱ | 86 | 0.156 (0.037, 0.543) | 0.133 | N3 | 9 | 0.374 (0.148, 0.576) | 0.306 | |
| Ⅲ | 14 | 0.055 (0.017, 0.231) | 0.382 |
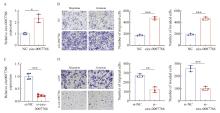
Fig. 3
Circ-0007766 promotes breast cancer cell invasion and migration in vitro A: The level of circ-0007766 in MDA-MB-231 cells was detected by RTFQ-PCR after transfection with circ-0007766 or NC plasmid. B: The number of migrating and invasive MDA-MB-231 cells was shown by transwell assay after transfection with circ-0007766 or NC plasmid. C: The expression of circ-0007766 in SK-BR-3 cells after transfection with si-circ-0007766 or si-NC plasmid. D: The number of migrating and invasive cells in SK-BR-3 cells after transfection with si-NC and si-circ-0007766 plasmid was shown by transwell assay. *: P<0.05, compared with NC; **: P<0.01, compared with si-NC; ***: P<0.001, compared with NC or si-NC."
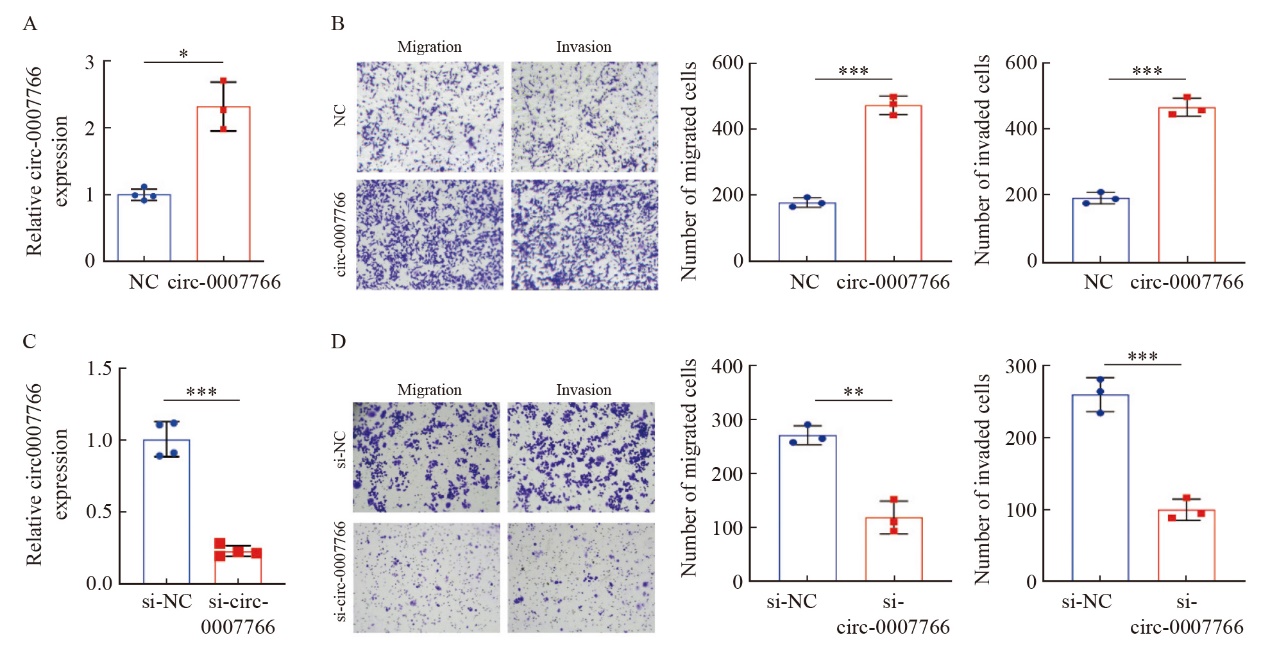
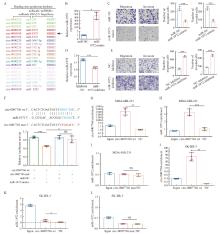
Fig. 4
Circ-0007766 interacts with miR-1972, a tumor suppressor gene A: The schematic diagram showed the predicted circR-NA-miRNA-HER2 regulatory network. CircRNAs, miRNAs, and HER2 on the same regulatory axis were identified by the same color. B: The expression of miR-1972 in SK-BR-3 cells was evaluated after transfection with NC mimics (miR-NC) or miR-1972 mimic. C: The migration and invasion ability of SK-BR-3 cells was determined by transwell assay after transfection with miR-NC or miR-1972 mimic. D: The level of miR-1972 in MDA-MB-231 cells was determined after transfection with miR-1972 inhibitor or inhibitor NC. E: The migration and invasion ability of MDA-MB-231 cells was determined by transwell assay after transfection with miR-1972 inhibitor or inhibitor NC. F: The top of the figure described the predicted binding site between circ-0007766 and miR-1972, as well as the mutation strategy to change the binding site between circ-0007766 and miR-1972. The relative luciferase activity of miR-1972 mimics, miR-NC, circ-0007766-3'UTR-wt, or circ-0007766-3'UTR-mut co-transfected with 293T cells was shown in the figure below. G-I: The RAP assay and RTFQ-PCR assay detected the enrichment of circ-0007766 wt and circ-0007766 mut probes for circ-0007766 and miR-1972 in MDA-MB-231 cells. J-L: The RAP assay and RTFQ-PCR assay detected the enrichment of circ-0007766 wt and circ-0007766 mut probes for circ-0007766 and miR-1972 in MDA-MB-231 cells. *: P <0.05, compared with circ-0007766 mut or miR-NC; **: P <0.01, compared with circ-0007766 wt; ***: P<0.001, compared with Inhibitor-NC, miR-NC, NC inhibitor or circ-0007766 wt; NS: No significant difference. wt: Wild type; mut: Mutant."
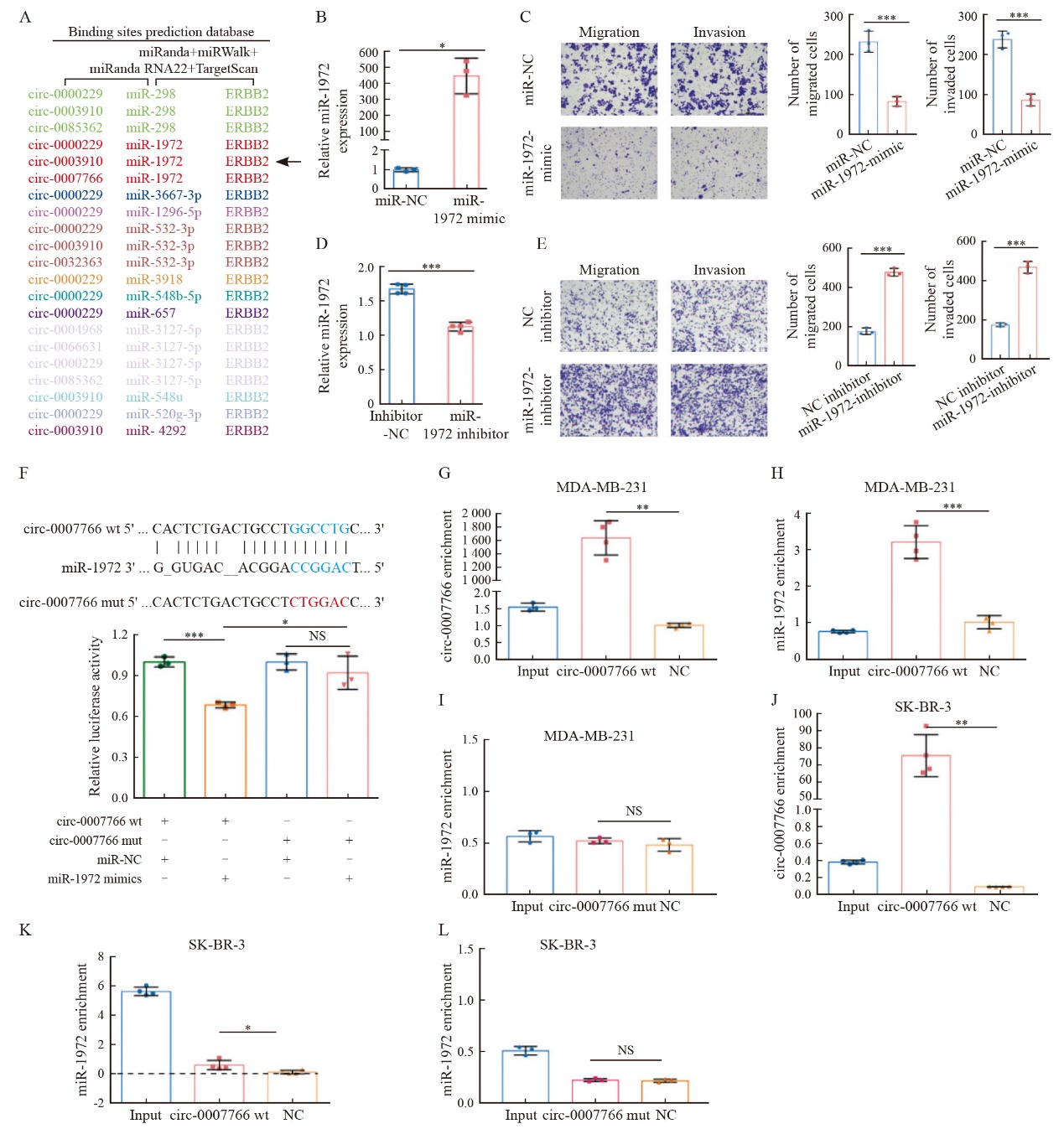
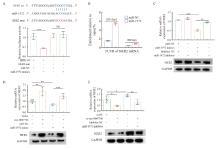
Fig. 5
MiR-1972 binds to HER2 mRNA and reverses the regulatory effect of circ-0007766 on HER2 expression A: The top of the figure showed the predetermined binding sites of miR-1972 on HER2 mRNA and the mutation strategies for the binding sites of HER2 mRNA with miR-1972. The bottom of the figure showed the relative luciferase activity in 293T cells after co-transfection with miR-1972 mimics, miR-NC, HER2-3'UTR-mut (HER2-mut) or HER2-3'UTR-wt (HER2-wt). B: After co-transfection with miR-NC or miR-1972 mimics, RIP assay was performed in MDA-MB-231 cells, and HER2 mRNA 3'UTR was measured by RTFQ-PCR. C: RTFQ-PCR and Western blot confirmed the expression levels of HER2 mRNA and protein in MDA-MB-231 cells after co-transfection with miR-NC+NC, miR-NC+miR-1972, miR-1972+NC or their combinations, and specific inhibitors. D: RTFQ-PCR and Western blot showed the expression levels of HER2 mRNA and protein in MDA-MB-231 cells after co-transfection with vector+miR-NC, circ-0007766+miR-NC, vector+miR-1972 mimics or circ-0007766+miR-1972 mimics. E: RTFQ-PCR and Western blot showed the expression levels of HER mRNA and protein in SK-BR-3 cells after co-transfection with si-NC+inhibitor NC, si-circ-0007766+inhibitor NC, si-NC+miR-1972 inhibitor or si-circ-0007766+miR-1972 inhibitor. *: P<0.05, compared with si-NC or inhibitor NC; **: P<0.01, compared with inhibitor NC, vector, miR-NC or si-NC; ***: P<0.001, compared with miR-NC or vector; NS: No significant difference."
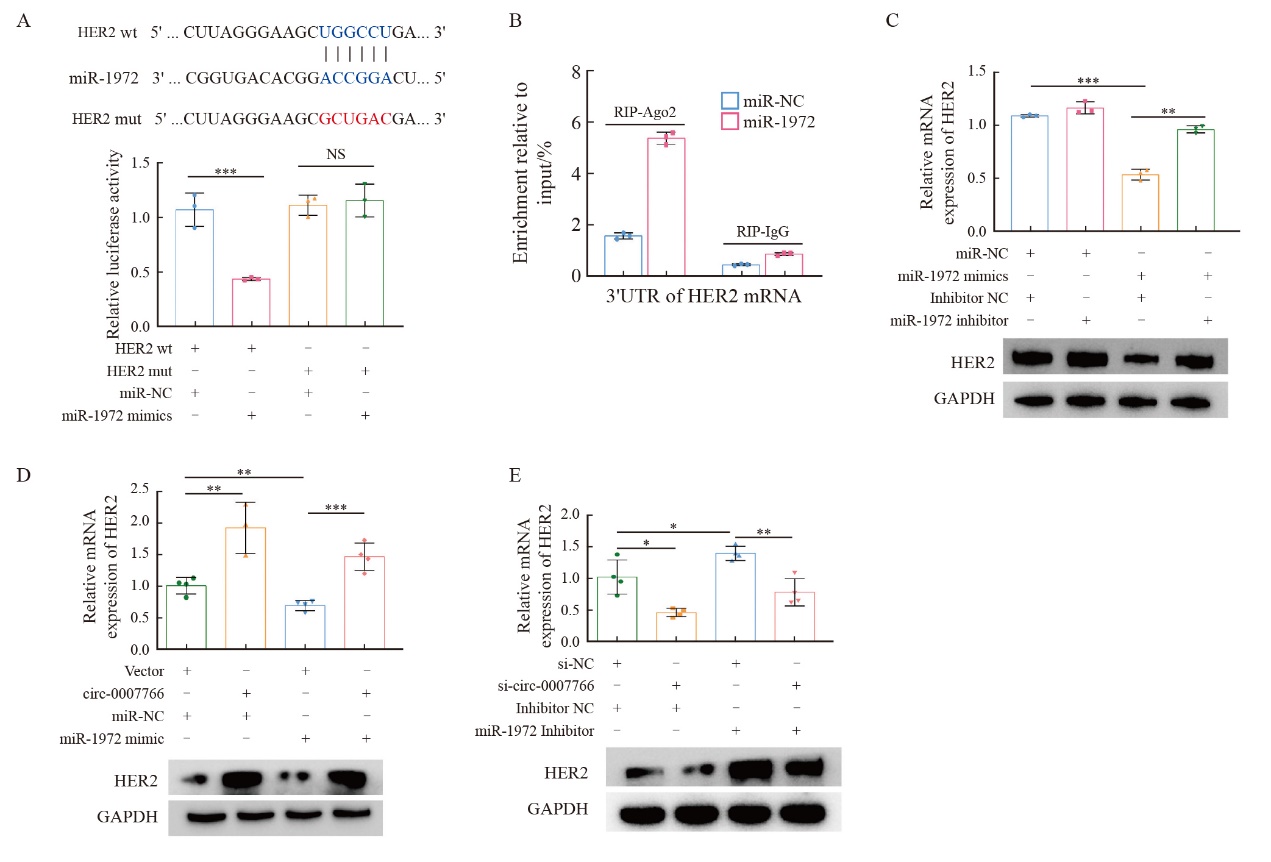
Fig. 6
Circ-0007766 promotes breast cancer cell migration and invasion through miR-1972 A: The migration and invasion potential of MDA-MB-231 cells transfected with the transfection vector+miR-NC, circ-0007766+miR-NC, vector+miR-1972 mimic or circ-0007766+miR-1972 mimic was evaluated using a transwell migration and invasion assay. B: The migration and invasion ability of SK-BR-3 cells after transfection with si-NC+inhibitors NC, si-circ-0007766+inhibitors NC, si-NC+miR-1972 inhibitor or si-circ-0007766+miR-1972 inhibitor was evaluated using a transwell assay. *: P<0.05, compared with si-NC; **: P<0.01, compared with vector, miR-NC, si-NC or inhibitor NC; ***: P<0.001, compared with vector, inhibitor NC or si-NC."
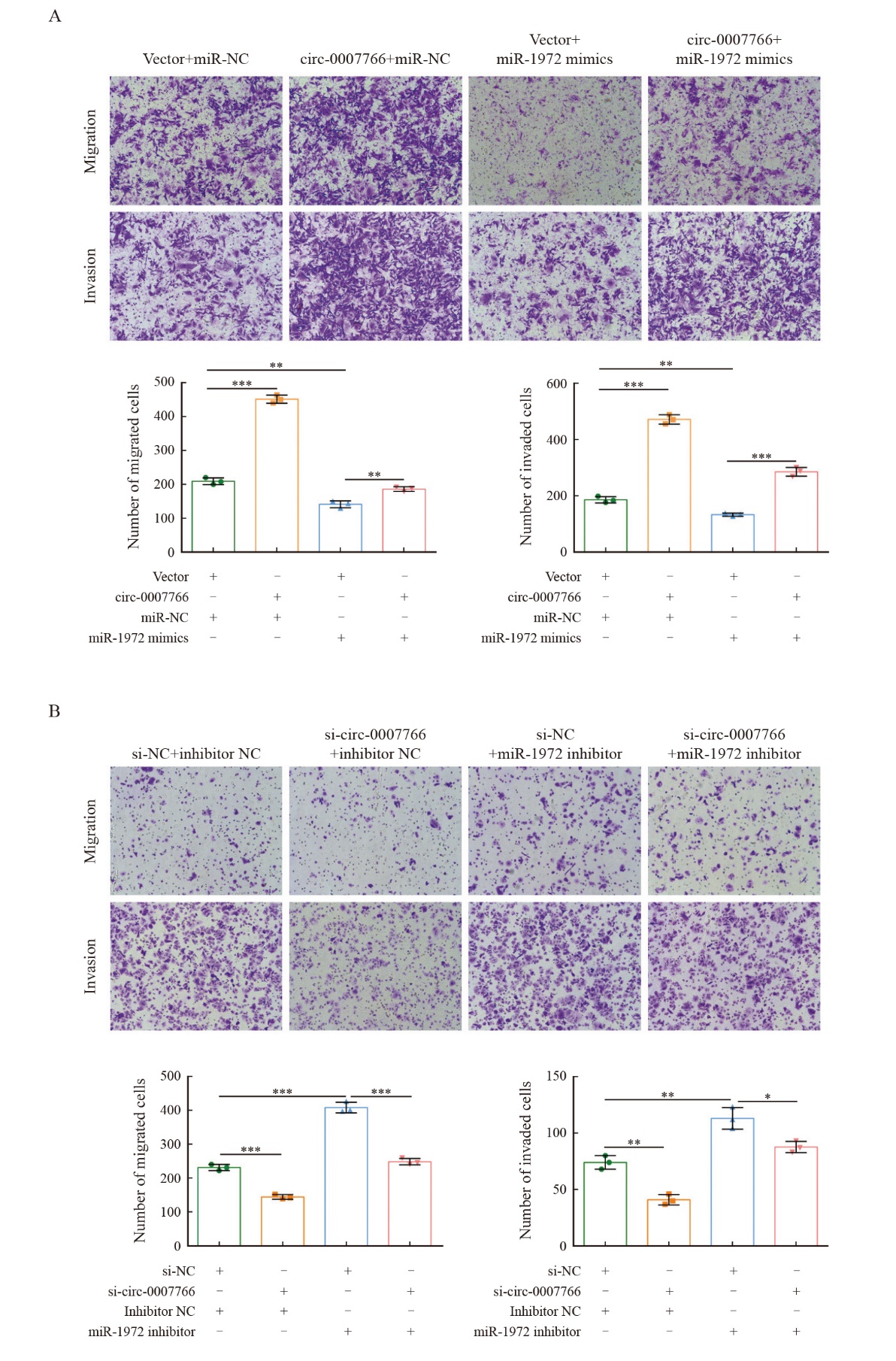
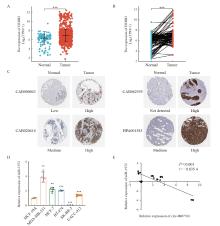
Fig. 7
Expression of HER2 in breast cancer tissues and correlation of circ-0007766 and miR-1972 expression in breast cancer cells A: Expression of HER2 mRNA in unpaired TCGA breast cancer/normal tissue. B: Expression of HER2 mRNA in paired TCGA-derived breast cancer tissue and normal tissue. C: Expression of HER2 protein in breast cancer and normal tissue, shown by representative immunohistochemical staining images from the Human Protein Atlas. D: Expression of miR-1972 in different breast cancer cells and normal breast cancer cell MCF10A. E: Analysis of the correlation between circ-0007766 and miR-1972 expression using data from Fig. 1C and Fig. 6D. Circ-0007766 and miR-1972 mRNA relative expression levels were converted to log2 scale. **: P<0.01, compared with MCF10A; ***: P<0.001, compared with normal or MCF10A."
| [1] |
YEO S K, GUAN J L. Breast cancer: multiple subtypes within a tumor?[J]. Trends Cancer, 2017, 3(11): 753-760.
doi: S2405-8033(17)30175-9 pmid: 29120751 |
| [2] | SUNG H, FERLAY J, SIEGEL R L, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71(3): 209-249. |
| [3] | LIU Y, ZHU X Z, XIAO Y, et al. Subtyping-based platform guides precision medicine for heavily pretreated metastatic triple-negative breast cancer: the FUTURE phase Ⅱ umbrella clinical trial[J]. Cell Res, 2023, 33(5): 389-402. |
| [4] |
HARBECK N, GNANT M. Breast cancer l[J]. Lancet, 2017, 389(10074): 1134-1150.
doi: S0140-6736(16)31891-8 pmid: 27865536 |
| [5] | OH D Y, BANG Y J. HER2-targeted therapies-a role beyond breast cancer[J]. Nat Rev Clin Oncol, 2020, 17: 33-48. |
| [6] | TAPIA M, HERNANDO C, MARTÍNEZ M T, et al. Clinical impact of new treatment strategies for HER2-positive metastatic breast cancer patients with resistance to classical anti-HER therapies[J]. Cancers, 2023, 15(18): 4522. |
| [7] | YE P, WANG Y R, LI R Q, et al. The HER family as therapeutic targets in colorectal cancer[J]. Crit Rev Oncol Hematol, 2022, 174: 103681. |
| [8] | SCERRI J, SCERRI C, SCHÄFER-RUOFF F, et al. PKC-mediated phosphorylation and activation of the MEK/ERK pathway as a mechanism of acquired trastuzumab resistance in HER2-positive breast cancer[J]. Front Endocrinol, 2022, 13: 1010092. |
| [9] | HARPER K L, SOSA M S, ENTENBERG D, et al. Mechanism of early dissemination and metastasis in HER2+ mammary cancer[J]. Nature, 2016, 540(7634): 588-592. |
| [10] |
LO P K, KANOJIA D, LIU X, et al. CD49f and CD61 identify HER2/neu-induced mammary tumor-initiating cells that are potentially derived from luminal progenitors and maintained by the integrin-TGFβ signaling[J]. Oncogene, 2012, 31(21): 2614-2626.
doi: 10.1038/onc.2011.439 pmid: 21996747 |
| [11] | NAMI B, WANG Z X. HER2 in breast cancer stemness: a negative feedback loop towards trastuzumab resistance[J]. Cancers, 2017, 9(5): 40. |
| [12] | LUO Z, RONG Z Y, ZHANG J M, et al. Circular RNA circCCDC9 acts as a miR-6792-3p sponge to suppress the progression of gastric cancer through regulating CAV1 expression[J]. Mol Cancer, 2020, 19(1): 86. |
| [13] | WU Y Z, XIE Z A, CHEN J X, et al. Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR-203a-3p and regulating CREB3 expression[J]. Mol Cancer, 2019, 18(1): 73. |
| [14] | LI J, MA M G, YANG X S, et al. Circular HER2 RNA positive triple negative breast cancer is sensitive to pertuzumab[J]. Mol Cancer, 2020, 19(1): 142. |
| [15] | 张帅, 夏文佳, 董高超, 等. 环状RNA分子circ_0007766通过上调细胞周期相关蛋白cyclin D1/cyclin E1/CDK4的表达促进肺腺癌细胞增殖[J]. 中国肺癌杂志, 2019, 22(5): 271-279. |
| ZHANG S, XIA W J, DONG G C, et al. Cyclic RNA molecule circ_0007766 promotes the proliferation of lung adenocarcinoma cells by up-regulating the expression of cyclin D1/cyclin E1/CDK4[J]. Chin J Lung Cancer, 2019, 22(5): 271-279. | |
| [16] | XU W G, ZHOU B, WU J, et al. Circular RNA hsa-circ-0007766 modulates the progression of Gastric Carcinoma via miR-1233-3p/GDF15 axis[J]. Int J Med Sci, 2020, 17(11): 1569-1583. |
| [17] | GUO J, PAN H. Long non-coding RNA LINC01125 enhances cisplatin sensitivity of ovarian cancer via miR-1972[J]. Med Sci Monit, 2019, 25: 9844-9854. |
| [18] | WANG S G, QIU J G, WANG L P, et al. Long non-coding RNA LINC01207 promotes prostate cancer progression by downregulating microRNA-1972 and upregulating LIM and SH3 protein 1[J]. IUBMB Life, 2020, 72(9): 1960-1975. |
| [19] | WANG Y, ZENG X D, WANG N N, et al. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma[J]. Mol Cancer, 2018, 17(1): 89. |
| [20] |
朱艺, 肖斌, 刘嘉慧, 等. Circ-0003910在HER2阳性乳腺癌中的表达、定位、生物学作用及蛋白质组学研究[J]. 中国癌症杂志, 2022, 32 (10): 979-989.
doi: 10.19401/j.cnki.1007-3639.2022.10.006 |
| ZHU Y, XIAO B, LIU J H, et al. Expression, localization, biological role and proteomics study of circ-0003910 in HER2-positive breast cancer[J]. China Oncol, 2022, 32(10): 979-989. | |
| [21] |
HUANG L, MA J, CUI M. Circular RNA hsa_circ_0001598 promotes programmed death-ligand-1-mediated immune escape and trastuzumab resistance via sponging miR-1184 in breast cancer cells[J]. Immunol Res, 2021, 69(6): 558-567.
doi: 10.1007/s12026-021-09237-w pmid: 34559381 |
| [22] | HOSONAGA M, ARIMA Y, SUGIHARA E, et al. Effect of heterogeneity of HER2 expression on brain metastases of breast cancer[J]. J Clin Oncol, 2012, 30(15_suppl): 635. |
| [23] | JORDAN N V, BARDIA A, WITTNER B S, et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells[J]. Nature, 2016, 537: 102-106. |
| [24] |
JIN J, CAO J, LI B, et al. Landscape of DNA damage response gene alterations in breast cancer: a comprehensive investigation[J]. Cancer, 2023, 129(6): 845-859.
doi: 10.1002/cncr.34618 pmid: 36655350 |
| [25] | ZHANG P, ZHANG Q Y, TONG Z S, et al. Dalpiciclib plus letrozole or anastrozole versus placebo plus letrozole or anastrozole as first-line treatment in patients with hormone receptor-positive, HER2-negative advanced breast cancer (DAWNA-2): a multicentre, randomised, double-blind, placebo-controlled, phase 3 trial[J]. Lancet Oncol, 2023, 24(6): 646-657. |
| [26] |
中国抗癌协会乳腺癌专业委员会,中华医学会肿瘤学分会乳腺肿瘤学组. 中国抗癌协会乳腺癌诊治指南与规范(2024年版)[J]. 中国癌症杂志, 2023, 33(12): 1092-1187.
doi: 10.19401/j.cnki.1007-3639.2023.12.004 |
| The Society of Breast Cancer China Anti-Cancer Association, Breast Oncology Group of the Oncology Branch of the Chinese Medical Association. Guidelines for breast cancer diagnosis and treatment by China Anti-Cancer Association (2024 edition)[J]. China Oncol, 2023, 33(12): 1092-1187. | |
| [27] |
ZHENG Q P, BAO C Y, GUO W J, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs[J]. Nat Commun, 2016, 7: 11215.
doi: 10.1038/ncomms11215 pmid: 27050392 |
| [28] |
KRISTENSEN L S, ANDERSEN M S, STAGSTED L V W, et al. The biogenesis, biology and characterization of circular RNAs[J]. Nat Rev Genet, 2019, 20(11): 675-691.
doi: 10.1038/s41576-019-0158-7 pmid: 31395983 |
| [29] | WANG Q, WANG H Z, ZHAO X M, et al. Transcriptome sequencing of circular RNA reveals the involvement of hsa-SCMH1_0001 in the pathogenesis of Parkinson’s disease[J]. CNS Neurosci Ther, 2024, 30(3): e14435. |
| [30] | LIU B Y, GONG Y J, JIANG Q Y, et al. Hsa_circ_0014784-induced YAP1 promoted the progression of pancreatic cancer by sponging miR-214-3p[J]. Cell Cycle, 2023, 22(13): 1583-1596. |
| [31] | WANG X S, XING L, YANG R, et al. The circACTN4 interacts with FUBP1 to promote tumorigenesis and progression of breast cancer by regulating the expression of proto-oncogene MYC[J]. Mol Cancer, 2021, 20(1): 91. |
| [32] | CAO L L, WANG M, DONG Y J, et al. Circular RNA circRNF20 promotes breast cancer tumorigenesis and Warburg effect through miR-487a/HIF-1α/HK2[J]. Cell Death Dis, 2020, 11(2): 145. |
| [33] | GUO X Y, HE C X, WANG Y Q, et al. Circular RNA profiling and bioinformatic modeling identify its regulatory role in hepatic steatosis[J]. Biomed Res Int, 2017, 2017: 5936171. |
| [34] | KIRBY E, TSE W H, PATEL D, et al. First steps in the development of a liquid biopsy in situ hybridization protocol to determine circular RNA biomarkers in rat biofluids[J]. Pediatr Surg Int, 2019, 35(12): 1329-1338. |
| [1] | CHEN Yijun, LIU Yuhang, DUAN Haibo, WANG Xiongjun. Functional and mechanistic of AGPAT5 in liver cancer [J]. China Oncology, 2024, 34(9): 838-847. |
| [2] | XU Rui, WANG Zehao, WU Jiong. Advances in the role of tumor-associated neutrophils in the development of breast cancer [J]. China Oncology, 2024, 34(9): 881-889. |
| [3] | CAO Xiaoshan, YANG Beibei, CONG Binbin, LIU Hong. The progress of treatment for brain metastases of triple-negative breast cancer [J]. China Oncology, 2024, 34(8): 777-784. |
| [4] | ZHANG Jian. Clinical consideration of two key questions in assessing menopausal status of female breast cancer patients [J]. China Oncology, 2024, 34(7): 619-627. |
| [5] | JIANG Dan, SONG Guoqing, WANG Xiaodan. Study on the mechanism of mitochondrial dysfunction and CPT1A/ERK signal transduction pathway regulating malignant behavior in breast cancer [J]. China Oncology, 2024, 34(7): 650-658. |
| [6] | HUANG Sijie, KANG Xun, LI Wenbin. Clinical research progress of intrathecal therapy in the treatment of leptomeningeal metastasis [J]. China Oncology, 2024, 34(7): 695-701. |
| [7] | DONG Jianqiao, LI Kunyan, LI Jing, WANG Bin, WANG Yanhong, JIA Hongyan. A study on mechanism of SIRT3 inducing endocrine drug resistance in breast cancer via deacetylating YME1L1 [J]. China Oncology, 2024, 34(6): 537-547. |
| [8] | HAO Xian, HUANG Jianjun, YANG Wenxiu, LIU Jinting, ZHANG Junhong, LUO Yubei, LI Qing, WANG Dahong, GAO Yuwei, TAN Fuyun, BO Li, ZHENG Yu, WANG Rong, FENG Jianglong, LI Jing, ZHAO Chunhua, DOU Xiaowei. Establishment of primary breast cancer cell line as new model for drug screening and basic research [J]. China Oncology, 2024, 34(6): 561-570. |
| [9] | SHEN Jie, FENG Xiaoshuang, WEN Hao, ZHOU Changming, MO Miao, WANG Zezhou, YUAN Jing, WU Xiaohua, ZHENG Ying. Metastasis patterns and survival analysis of 572 patients with metastatic cervical cancer: a hospital-based real world study [J]. China Oncology, 2024, 34(4): 361-367. |
| [10] | LI Xiaohui, ZHAO Jiaxu, PENG Haibao, ZHANG Ye, ZENG Rui, CHI Yudan. Effects of HMGA2 on migration and proliferation of leptomeningeal metastatic melanoma [J]. China Oncology, 2024, 34(4): 389-399. |
| [11] | Committee of Breast Cancer Society, China Anti-Cancer Association. Expert consensus on clinical applications of ovarian function suppression for Chinese women with early breast cancer (2024 edition) [J]. China Oncology, 2024, 34(3): 316-333. |
| [12] | ZHANG Qi, XIU Bingqiu, WU Jiong. Progress of important clinical research of breast cancer in China in 2023 [J]. China Oncology, 2024, 34(2): 135-142. |
| [13] | ZHANG Siyuan, JIANG Zefei. Important research progress in clinical practice for advanced breast cancer in 2023 [J]. China Oncology, 2024, 34(2): 143-150. |
| [14] | WANG Zhaobu, LI Xing, YU Xinmiao, JIN Feng. Important research progress in clinical practice for early breast cancer in 2023 [J]. China Oncology, 2024, 34(2): 151-160. |
| [15] | LUO Yang, SUN Tao, SHAO Zhimin, CUI Jiuwei, PAN Yueyin, ZHANG Qingyuan, CHENG Ying, LI huiping, YANG Yan, YE Changsheng, YU Guohua, WANG Jingfen, LIU Yunjiang, LIU Xinlan, ZHOU Yuhong, BAI Yuju, GU Yuanting, WANG Xiaojia, XU Binghe, SONG Lihua. Efficacy, metabolic characteristics, safety and immunogenicity of AK-HER2 compared with reference trastuzumab in patients with metastatic HER2-positive breast cancer: a multicenter, randomized, double-blind phase Ⅲ equivalence trial [J]. China Oncology, 2024, 34(2): 161-175. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
沪ICP备12009617
Powered by Beijing Magtech Co. Ltd
