Welcome to China Oncology,
China Oncology ›› 2022, Vol. 32 ›› Issue (10): 979-989.doi: 10.19401/j.cnki.1007-3639.2022.10.006
• Article • Previous Articles Next Articles
ZHU Yi1,3( ), XIAO Bin1, LIU Jiahui2, HUANG Ling2, SUN Zhaohui2, LI Linhai1
), XIAO Bin1, LIU Jiahui2, HUANG Ling2, SUN Zhaohui2, LI Linhai1
Received:2022-03-24
Revised:2022-08-04
Online:2022-10-30
Published:2022-11-29
Share article
CLC Number:
ZHU Yi, XIAO Bin, LIU Jiahui, HUANG Ling, SUN Zhaohui, LI Linhai. Expression, localization, biological role and proteomics study of circ-0003910 in HER2-positive breast cancer[J]. China Oncology, 2022, 32(10): 979-989.
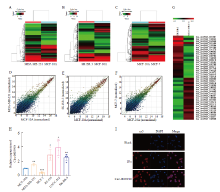
Fig. 2
Circ-0003910 was up-regulated in HER2 positive breast cancer cells A-C: Heat map of differentially expressed circRNA; D-F: Scatter plots of differentially expressed circRNA; G: Heat map of the top 20 circRNA with the highest differential expression; H: Expression of circ-0003910 in five types of breast cancer cells and normal breast cells; I: Subcellular localization of circ-0003910 in SK-BR-3 cells; *: P<0.05, **: P<0.01, ***: P<0.001, compared with MCF-10A."
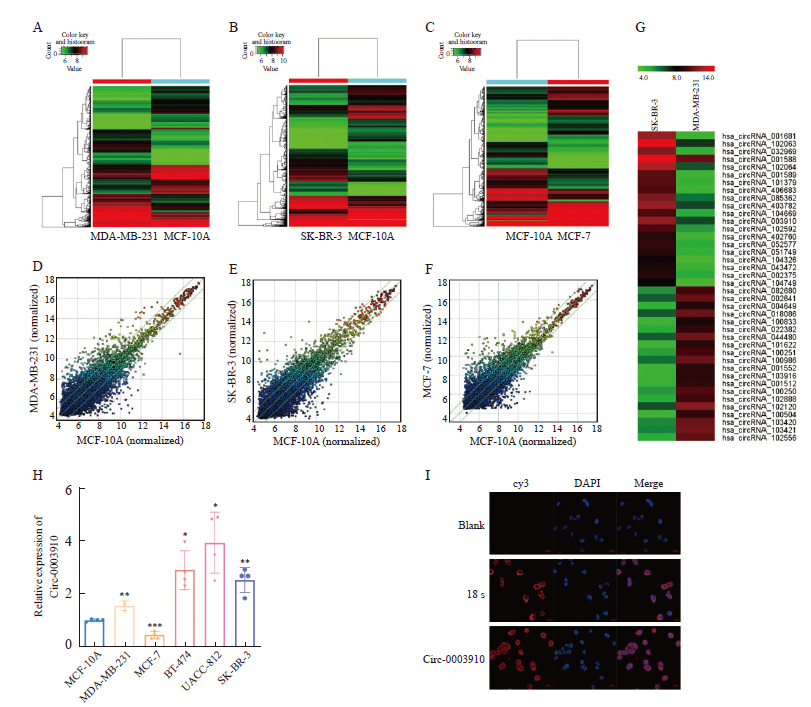
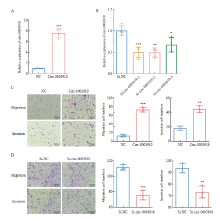
Fig. 4
Circ-0003910 promotes breast cancer cell migration and invasion in vitro A-B: The mRNA expression level of CIRC-0003910 was detected after overexpression and knockdown of circ-0003910 by RTFQ-PCR; C: Transwell assay detected the migration and invasion of MDA-MB-231 cells when circ-0003910 was overexpressed; D: Transwell assay detects the migration and invasion of SK-BR-3 cells when circ-0003910 was knocked down. **: P<0.01, compared with each other; ***: P<0.001, compared with each other."

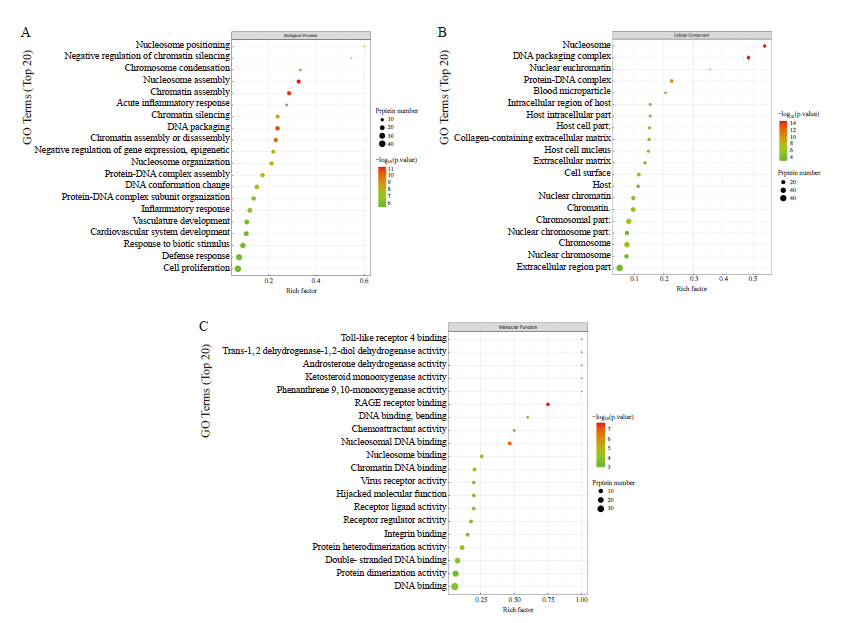
Fig. 5
GO enrichment analysis of downstream differentially expressed proteins affected by circ-0003910 overexpression A: GO analysis of differentially expressed proteins enriched in biological process; B: GO analysis of differentially expressed proteins enriched in cellular component; C: GO analysis of differentially expressed proteins enriched in molecular function"
| [1] |
SUNG H, FERLAY J, SIEGEL R L, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71(3): 209-249.
doi: 10.3322/caac.21660 |
| [2] |
YEO S K, GUAN J L. Breast cancer: multiple subtypes within a tumor?[J]. Trends Cancer, 2017, 3(11): 753-760.
doi: S2405-8033(17)30175-9 pmid: 29120751 |
| [3] |
HARBECK N, GNANT M. Breast cancer[J]. Lancet, 2017, 389(10074): 1134-1150.
doi: S0140-6736(16)31891-8 pmid: 27865536 |
| [4] | 王笑. HER2阳性乳腺癌中KIF3B表达意义及其与上皮间质转化的相关性研究[D]. 青岛: 青岛大学, 2021. |
| WANG X. Significance of KIF3B expression in HER2 positive breast cancer and its correlation with epithelial mesenchymal transition[D] Qingdao: Qingdao University, 2021. | |
| [5] |
ZHENG Q P, BAO C Y, GUO W J, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs[J]. Nat Commun, 2016, 7: 11215.
doi: 10.1038/ncomms11215 pmid: 27050392 |
| [6] |
YIN Y T, LONG J L, HE Q L, et al. Emerging roles of circRNA in formation and progression of cancer[J]. J Cancer, 2019, 10(21): 5015-5021.
doi: 10.7150/jca.30828 pmid: 31602252 |
| [7] |
LIN X S, HUANG C, CHEN Z A, et al. CircRNA_100876 is upregulated in gastric cancer (GC) and promotes the GC cells' growth, migration and invasion via miR-665/YAP1 signaling[J]. Front Genet, 2020, 11: 546275.
doi: 10.3389/fgene.2020.546275 |
| [8] |
LIU H L, LAN T, LI H, et al. Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR[J]. Theranostics, 2021, 11(3): 1396-1411.
doi: 10.7150/thno.53227 pmid: 33391541 |
| [9] |
CHEN D S, MA W, KE Z Y, et al. CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression[J]. Cell Cycle, 2018, 17(16): 2080-2090.
doi: 10.1080/15384101.2018.1515553 pmid: 30176158 |
| [10] |
LIU Z H, ZHOU Y, LIANG G H, et al. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p[J]. Cell Death Dis, 2019, 10(2): 55.
doi: 10.1038/s41419-018-1287-1 pmid: 30670688 |
| [11] | 王晓松, 陈俊霞. 环状RNA hsa_circ_0050900在乳腺癌中的表达及其对细胞生物学行为影响的机制研究[J]. 中国癌症杂志, 2020, 30(12): 977-983. |
| WANG X S, CHEN J X. Expression of circular RNA has_circ_0050900 in breast cancer and its effect on cell function[J]. China Oncol, 2020, 30(12): 977-983. | |
| [12] |
LUO Z, RONG Z Y, ZHANG J M, et al. Circular RNA circCCDC9 acts as a miR-6792-3p sponge to suppress the progression of gastric cancer through regulating CAV1 expression[J]. Mol Cancer, 2020, 19(1): 86.
doi: 10.1186/s12943-020-01203-8 pmid: 32386516 |
| [13] |
XU J Z, SHAO C C, WANG X J, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis[J]. Cell Death Dis, 2019, 10(3): 175.
doi: 10.1038/s41419-019-1382-y |
| [14] | ZEESHAN R, MUTAHIR Z. Cancer metastasis-tricks of the trade[J]. Bosn J Basic Med Sci, 2017, 17(3): 172-182. |
| [15] |
LI T, MELLO-THOMS C, BRENNAN P C. Descriptive epidemiology of breast cancer in China: incidence, mortality, survival and prevalence[J]. Breast Cancer Res Treat, 2016, 159(3): 395-406.
doi: 10.1007/s10549-016-3947-0 |
| [16] |
ZANG J K, LU D, XU A D. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function[J]. J Neurosci Res, 2020, 98(1): 87-97.
doi: 10.1002/jnr.24356 pmid: 30575990 |
| [17] |
KRISTENSEN L S, ANDERSEN M S, STAGSTED L V W, et al. The biogenesis, biology and characterization of circular RNAs[J]. Nat Rev Genet, 2019, 20(11): 675-691.
doi: 10.1038/s41576-019-0158-7 pmid: 31395983 |
| [18] | KLINGE C M. Non-coding RNAs in breast cancer: Intracellular and intercellular communication[J]. Noncoding RNA, 2018, 4(4): 40. |
| [19] |
KIRBY E, TSE W H, PATEL D, et al. First steps in the development of a liquid biopsy in situ hybridization protocol to determine circular RNA biomarkers in rat biofluids[J]. Pediatr Surg Int, 2019, 35(12): 1329-1338.
doi: 10.1007/s00383-019-04558-2 |
| [20] |
HUANG A Q, ZHENG H X, WU Z Y, et al. Circular RNA-protein interactions: functions, mechanisms, and identification[J]. Theranostics, 2020, 10(8): 3503-3517.
doi: 10.7150/thno.42174 pmid: 32206104 |
| [21] |
HU C J, NI Z H, LI B S, et al. hTERT promotes the invasion of gastric cancer cells by enhancing FOXO3a ubiquitination and subsequent ITGB1 upregulation[J]. Gut, 2017, 66(1): 31-42.
doi: 10.1136/gutjnl-2015-309322 pmid: 26370108 |
| [22] |
REN L L, MO W J, WANG L L, et al. Matrine suppresses breast cancer metastasis by targeting ITGB1 and inhibiting epithelial-to-mesenchymal transition[J]. Exp Ther Med, 2020, 19(1): 367-374.
doi: 10.3892/etm.2019.8207 pmid: 31853313 |
| [23] |
TAFTAF R, LIU X, SINGH S, et al. ICAM1 initiates CTC cluster formation and trans-endothelial migration in lung metastasis of breast cancer[J]. Nat Commun, 2021, 12(1): 4867.
doi: 10.1038/s41467-021-25189-z pmid: 34381029 |
| [24] |
FIGENSCHAU S L, KNUTSEN E, URBAROVA I, et al. ICAM1 expression is induced by proinflammatory cytokines and associated with TLS formation in aggressive breast cancer subtypes[J]. Sci Rep, 2018, 8(1): 11720.
doi: 10.1038/s41598-018-29604-2 pmid: 30082828 |
| [25] |
ALTEVOGT P, DOBERSTEIN K, FOGEL M. L1CAM in human cancer[J]. Int J Cancer, 2016, 138(7): 1565-1576.
doi: 10.1002/ijc.29658 pmid: 26111503 |
| [26] |
LI Y, GALILEO D S. Soluble L1CAM promotes breast cancer cell adhesion and migration in vitro, but not invasion[J]. Cancer Cell Int, 2010, 10: 34.
doi: 10.1186/1475-2867-10-34 |
| [1] | XU Rui, WANG Zehao, WU Jiong. Advances in the role of tumor-associated neutrophils in the development of breast cancer [J]. China Oncology, 2024, 34(9): 881-889. |
| [2] | CAO Xiaoshan, YANG Beibei, CONG Binbin, LIU Hong. The progress of treatment for brain metastases of triple-negative breast cancer [J]. China Oncology, 2024, 34(8): 777-784. |
| [3] | ZHANG Jian. Clinical consideration of two key questions in assessing menopausal status of female breast cancer patients [J]. China Oncology, 2024, 34(7): 619-627. |
| [4] | JIANG Dan, SONG Guoqing, WANG Xiaodan. Study on the mechanism of mitochondrial dysfunction and CPT1A/ERK signal transduction pathway regulating malignant behavior in breast cancer [J]. China Oncology, 2024, 34(7): 650-658. |
| [5] | DONG Jianqiao, LI Kunyan, LI Jing, WANG Bin, WANG Yanhong, JIA Hongyan. A study on mechanism of SIRT3 inducing endocrine drug resistance in breast cancer via deacetylating YME1L1 [J]. China Oncology, 2024, 34(6): 537-547. |
| [6] | HAO Xian, HUANG Jianjun, YANG Wenxiu, LIU Jinting, ZHANG Junhong, LUO Yubei, LI Qing, WANG Dahong, GAO Yuwei, TAN Fuyun, BO Li, ZHENG Yu, WANG Rong, FENG Jianglong, LI Jing, ZHAO Chunhua, DOU Xiaowei. Establishment of primary breast cancer cell line as new model for drug screening and basic research [J]. China Oncology, 2024, 34(6): 561-570. |
| [7] | Committee of Breast Cancer Society, China Anti-Cancer Association. Expert consensus on clinical applications of ovarian function suppression for Chinese women with early breast cancer (2024 edition) [J]. China Oncology, 2024, 34(3): 316-333. |
| [8] | ZHANG Qi, XIU Bingqiu, WU Jiong. Progress of important clinical research of breast cancer in China in 2023 [J]. China Oncology, 2024, 34(2): 135-142. |
| [9] | ZHANG Siyuan, JIANG Zefei. Important research progress in clinical practice for advanced breast cancer in 2023 [J]. China Oncology, 2024, 34(2): 143-150. |
| [10] | WANG Zhaobu, LI Xing, YU Xinmiao, JIN Feng. Important research progress in clinical practice for early breast cancer in 2023 [J]. China Oncology, 2024, 34(2): 151-160. |
| [11] | LUO Yang, SUN Tao, SHAO Zhimin, CUI Jiuwei, PAN Yueyin, ZHANG Qingyuan, CHENG Ying, LI huiping, YANG Yan, YE Changsheng, YU Guohua, WANG Jingfen, LIU Yunjiang, LIU Xinlan, ZHOU Yuhong, BAI Yuju, GU Yuanting, WANG Xiaojia, XU Binghe, SONG Lihua. Efficacy, metabolic characteristics, safety and immunogenicity of AK-HER2 compared with reference trastuzumab in patients with metastatic HER2-positive breast cancer: a multicenter, randomized, double-blind phase Ⅲ equivalence trial [J]. China Oncology, 2024, 34(2): 161-175. |
| [12] | CHEN Yuanxiang, YU Tao, YANG Shiyu, ZENG Tao, WEI Lan, ZHANG Yan. KDM4A promotes the migration and invasion of breast cancer cell line MDA-MB-231 by downregulating BMP9 [J]. China Oncology, 2024, 34(2): 176-184. |
| [13] | HU Xiaoyu, CAI Yuwen, YE Fugui, SHAO Zhimin, HU Weigang, YU Keda. Impact of BRCA1/2 germline mutation on the incidence of second primary cancer following postoperative radiotherapy in patients with triple-negative breast cancer [J]. China Oncology, 2024, 34(2): 185-190. |
| [14] | ZHANG Siwei, MA Ding, JIANG Yizhou, SHAO Zhimin. “Subtype-precise” therapy leads diagnostic and therapeutic innovations: a new pattern for precision treatment of breast cancer [J]. China Oncology, 2024, 34(11): 1045-1052. |
| [15] | OUYANG Fei, WANG Yang, CHEN Yu, PEI Guoqing, WANG Ling, ZHANG Yang, SHI Lei. Construction of the prediction model of breast cancer bone metastasis based on machine learning [J]. China Oncology, 2024, 34(10): 903-914. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
沪ICP备12009617
Powered by Beijing Magtech Co. Ltd
