Welcome to China Oncology,
China Oncology ›› 2023, Vol. 33 ›› Issue (7): 673-685.doi: 10.19401/j.cnki.1007-3639.2023.07.005
• Article • Previous Articles Next Articles
YE Junling1( ), ZHENG Xiaoying1, GUO Xinjian1, CHEN Ruihui1, YANG Liu1, GOU Xiaodan1, JIANG Hanmei2
), ZHENG Xiaoying1, GUO Xinjian1, CHEN Ruihui1, YANG Liu1, GOU Xiaodan1, JIANG Hanmei2
Received:2022-09-22
Revised:2023-01-03
Online:2023-07-30
Published:2023-08-10
Contact:
YE Junling
Share article
CLC Number:
YE Junling, ZHENG Xiaoying, GUO Xinjian, CHEN Ruihui, YANG Liu, GOU Xiaodan, JIANG Hanmei. A study on mechanism of lncRNA-mediated SNHG5/miR-26a-5p/MTDH signal axis promoting metastasis of colorectal cancer[J]. China Oncology, 2023, 33(7): 673-685.
Tab. 1
Primer sequence"
| Gene | Primer sequence (5'-3') |
|---|---|
| LncRNA SNHG5 | Forward: TACTGGCTGCGCACTTCG |
| Reverse: CAGTAAAAGGGGAACACCA | |
| miR-26a-5p | Forward: GCTCTGAACGTAGATCCGAAC |
| Reverse: GTGCAGGGTCCGAGGT | |
| MTDH | Forward: AGCAAAGCAGCCACCAGAG |
| Reverse: AGGAAATGATGCGGTTGTA | |
| U6 | Forward: CTCGCTTCGGCAGCACA |
| Reverse: AACGCTTCACGAATTTGCGT | |
| GAPDH | Forward: CTCTTCCAGCCTTCCTTCCT |
| Reverse: AGCACTGTGTTGGCGTACAG |
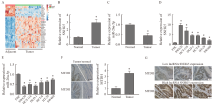
Fig. 1
Expression of lncRNA SNHG5, miR-26a-5p and MTDH in CRC A: TCGA database analysis results; B: Results of lncRNA SNHG5 RTFQ-PCR detection in CRC tissue; C: Results of miR-26a-5p RTFQ-PCR detection in CRC tissue; D: Results of lncRNA SNHG5 RTFQ-PCR detection of CRC cell line; E: Results of miR-26a-5p RTFQ-PCR detection of CRC cell line; F: Results of immunohistochemical detection of MTDH in CRC samples; G: Expression of MTDH in CRC tissues with different SNHG5 levels. *: P<0.05, compared with paracancerous tissues and FHC."
Tab. 2
Relationship between relative expression level of lncRNA SNHG5 in CRC and clinicopathological features"
| Characteristic | Case n (%) | LncRNA SNHG5 expression level $\bar{x}±s$ | t value | P value |
|---|---|---|---|---|
| Age/year | 0.630 | 0.530 | ||
| ≥60 | 56 (56.00) | 2.26±0.17 | ||
| <60 | 44 (44.00) | 2.24±0.14 | ||
| Gender | 0.292 | 0.771 | ||
| Male | 59 (59.00) | 2.25±0.18 | ||
| Female | 41 (41.00) | 2.24±0.15 | ||
| T stage | 2.885 | 0.005 | ||
| T1-2 | 69 (69.00) | 2.21±0.17 | ||
| T3-4 | 31 (31.00) | 2.32±0.19 | ||
| N stage | 2.908 | 0.004 | ||
| N0 | 79 (79.00) | 2.22±0.18 | ||
| N1 +N2 | 21 (21.00) | 2.35±0.19 | ||
| M stage | 2.839 | 0.005 | ||
| M0 | 80 (80.00) | 2.21±0.17 | ||
| M1 +M2 | 20 (20.00) | 2.34±0.23 | ||
| TNM stage | 3.823 | 0.000 | ||
| Ⅰ-Ⅱ | 73 (73.00) | 2.17±0.20 | ||
| Ⅲ | 27 (27.00) | 2.34±0.19 |
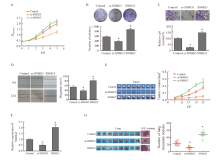
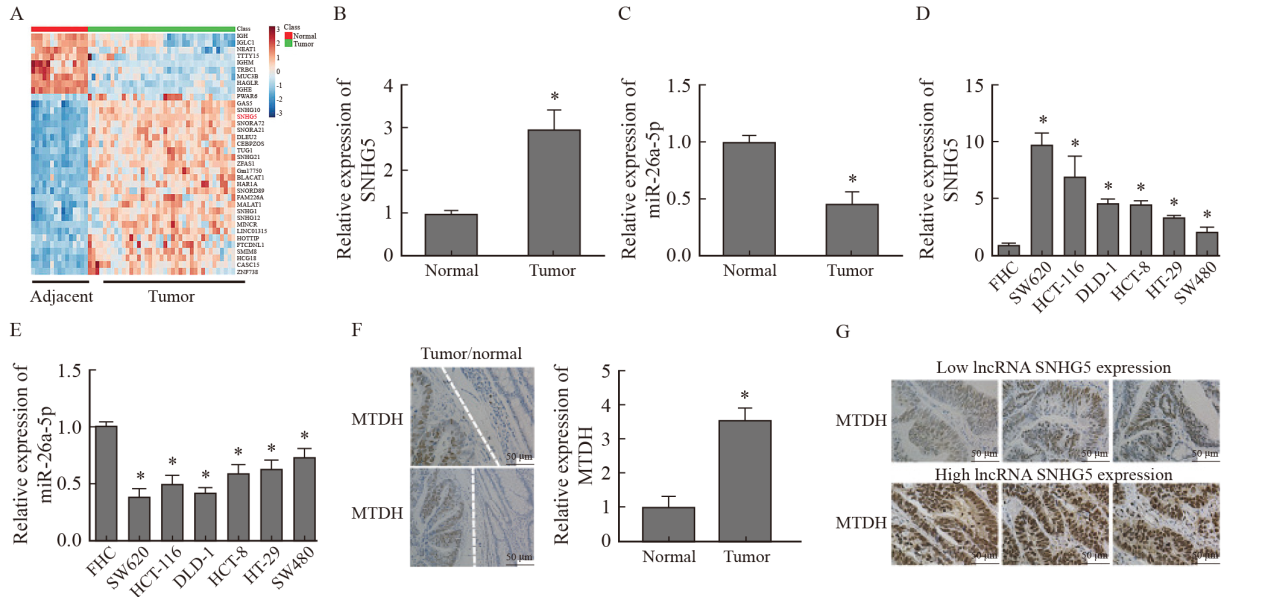
Fig. 3
Effects of lncRNA SNHG5 on proliferation, migration and invasion of CRC cells A: CCK-8 test results; B: Experimental results of clone formation; C: Transwell experimental results; D: Scratch test results; E: Experimental results of xenotransplantation in vivo; F: Results of lncRNA SNHG5 RTFQ-PCR detection of transplanted tumor; G: H-E staining results and number of metastatic pulmonary nodules. *: P<0.05, compared with control group."

Tab. 5
Comparison of relative expression levels of E-cadherin and vimentin in cells of different groups ($\bar{x}±s$)"
| Group | E-cadherin/GAPDH | Vimentin/GAPDH |
|---|---|---|
| Control group | 0.90±0.08 | 1.12±0.09 |
| si-SNHG5 group | 1.15±0.09 | 0.81±0.05 |
| SNHG5 group | 0.61±0.07 | 1.44±0.12 |
| F value | 122.283 | 116.203 |
| P value | 0.000 | 0.000 |
Tab. 6
Comparison of relative expression levels of E-cadherin, vimentin and Ki-67 proliferation index in different groups of transplanted tumors ($\bar{x}±s$)"
| Group | Ki-67 proliferation index | E-cadherin | Vimentin |
|---|---|---|---|
| Control group | 2.02±0.20 | 0.88±0.12 | 0.63±0.11 |
| si-SNHG5 group | 1.20±0.34 | 1.46±0.32 | 0.33±0.09 |
| SNHG5 group | 2.69±0.17 | 0.32±0.06 | 1.04±0.29 |
| F value | 78.102 | 65.896 | 29.520 |
| P value | 0.000 | 0.000 | 0.000 |
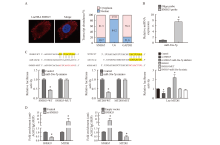
Fig. 5
Physical interaction between SNHG5 and miR-26a-5p, MTDH and miR-26a-5p A: Results of FISH detection; B: Results of RNA pull-down experiment; C: Detection results of double luciferase reporter gene; D: Results of co-immunoprecipitation. *: P<0.05, compared with control group. AGO2: Argonaute 2; IgG: immunoglobulin G; RIP: RNA immunoprecipitation."
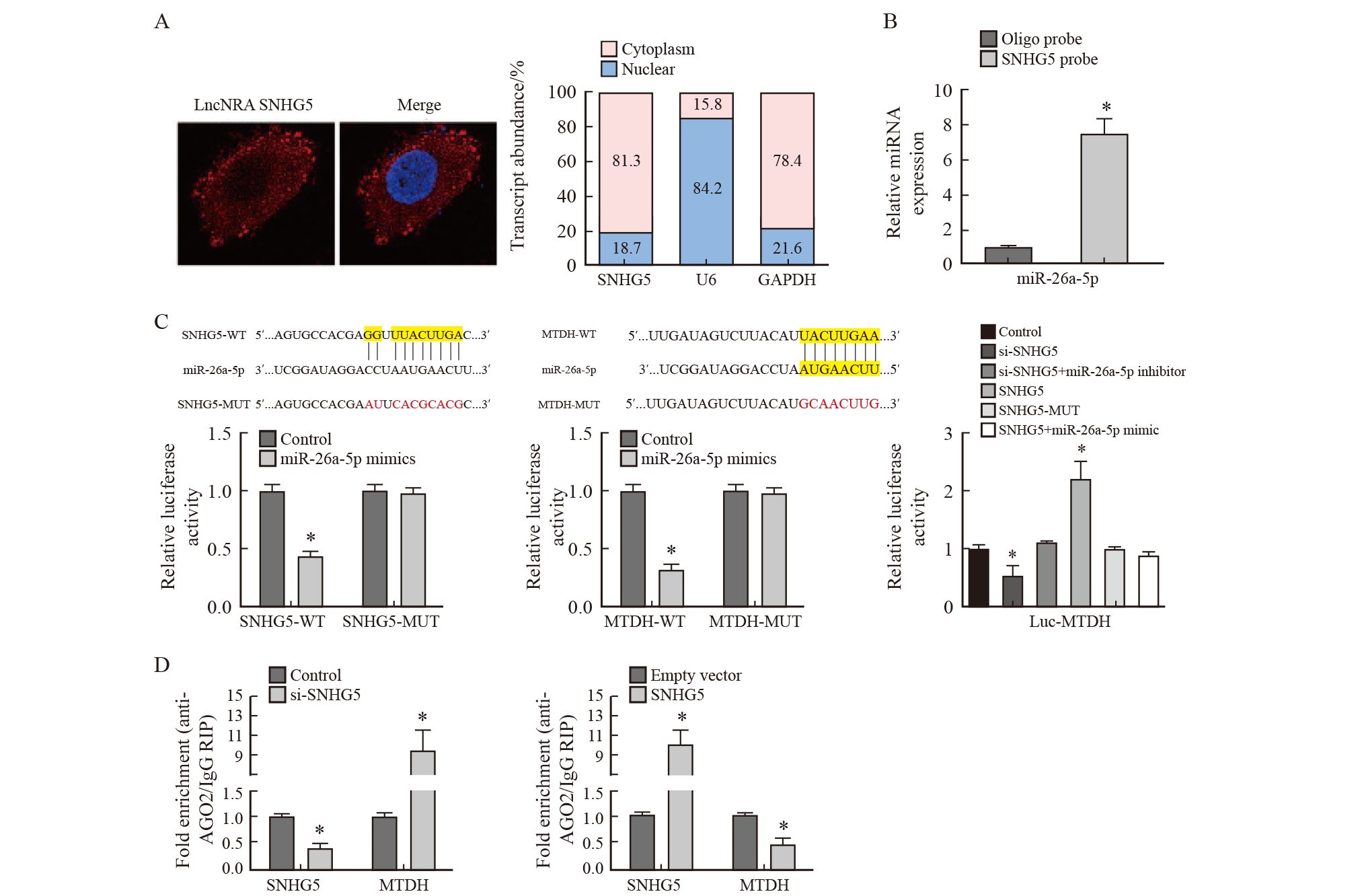
Tab. 7
Comparison of relative expression level of MTDH in cells of different groups"
| Group | MTDH/GAPDH $\bar{x}±s$ |
|---|---|
| Control group | 1.00±0.05 |
| si-SNHG5 group | 0.50±0.07 |
| si-SNHG5+miR-26a-5p inhibitor group | 0.99±0.05 |
| SNHG5 group | 1.72±0.14 |
| SNHG5+miR-26a-5p mimic group | 1.01±0.06 |
| F value | 289.479 |
| P value | 0.000 |
Tab. 8
Comparison of relative expression levels of MTDH, migration and invasion related molecules in different groups of cells ($\bar{x}±s$)"
| Group | E-cadherin/GAPDH | Vimentin/GAPDH | Cyclin D1/GAPDH | MMP-9/GAPDH | MTDH/GAPDH |
|---|---|---|---|---|---|
| Control group | 1.21±0.07 | 0.98±0.09 | 0.91±0.07 | 0.99±0.08 | 0.63±0.09 |
| si-SNHG5 group | 1.81±0.14 | 0.28±0.04 | 0.37±0.09 | 0.47±0.06 | 0.30±0.05 |
| SNHG5 group | 0.43±0.09 | 1.39±0.11 | 1.25±0.12 | 1.33±0.11 | 1.36±0.11 |
| si-MTDH group | 1.68±0.16 | 0.33±0.04 | 0.43±0.09 | 0.45±0.07 | 0.14±0.04 |
| SNHG5+si-MTDH group | 1.24±0.07 | 0.87±0.09 | 0.83±0.07 | 1.02±0.09 | 0.67±0.11 |
| F value | 245.203 | 443.520 | 196.911 | 187.544 | 289.031 |
| P value | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| [1] |
OGUNWOBI O O, MAHMOOD F, AKINGBOYE A. Biomarkers in colorectal cancer: current research and future prospects[J]. Int J Mol Sci, 2020, 21(15): 5311.
doi: 10.3390/ijms21155311 |
| [2] |
LADABAUM U, DOMINITZ J A, KAHI C, et al. Strategies for colorectal cancer screening[J]. Gastroenterology, 2020, 158(2): 418-432.
doi: S0016-5085(19)41185-2 pmid: 31394083 |
| [3] |
VACANTE M, CIUNI R, BASILE F, et al. The liquid biopsy in the management of colorectal cancer: an overview[J]. Biomedicines, 2020, 8(9): 308.
doi: 10.3390/biomedicines8090308 |
| [4] |
SINGH N, EBERHARDT M, WOLKENHAUER O, et al. An integrative network-driven pipeline for systematic identification of lncRNA-associated regulatory network motifs in metastatic melanoma[J]. BMC Bioinformatics, 2020, 21(1): 329.
doi: 10.1186/s12859-020-03656-6 pmid: 32703153 |
| [5] |
WEI S S, SUN S P, ZHOU X L, et al. SNHG5 inhibits the progression of EMT through the ubiquitin-degradation of MTA2 in oesophageal cancer[J]. Carcinogenesis, 2021, 42(2): 315-326.
doi: 10.1093/carcin/bgaa110 pmid: 33095847 |
| [6] |
HUANG S L, HUANG Z C, ZHANG C J, et al. LncRNA SNHG5 promotes the glycolysis and proliferation of breast cancer cell through regulating BACH1 via targeting miR-299[J]. Breast Cancer, 2022, 29(1): 65-76.
doi: 10.1007/s12282-021-01281-6 |
| [7] |
WANG R Q, LONG X R, ZHOU N N, et al. Lnc-GAN1 expression is associated with good survival and suppresses tumor progression by sponging miR-26a-5p to activate PTEN signaling in non-small cell lung cancer[J]. J Exp Clin Cancer Res, 2021, 40(1): 9.
doi: 10.1186/s13046-020-01819-0 |
| [8] |
CAI Q, WANG C, HUANG L, et al. Long non-coding RNA small nucleolar RNA host gene 5 (SNHG5) regulates renal tubular damage in diabetic nephropathy via targeting miR-26a-5p[J]. Horm Metab Res, 2021, 53(12): 818-824.
doi: 10.1055/a-1678-6556 pmid: 34891212 |
| [9] |
SHEN M H, XIE S S, ROWICKI M, et al. Therapeutic targeting of metadherin suppresses colorectal and lung cancer progression and metastasis[J]. Cancer Res, 2021, 81(4): 1014-1025.
doi: 10.1158/0008-5472.CAN-20-1876 pmid: 33239430 |
| [10] | 中华人民共和国国家卫生健康委员会医政医管局, 中华医学会肿瘤学分会. 中国结直肠癌诊疗规范(2020年版)[J]. 中华外科杂志, 2020, 58(8): 601-625. |
| Hospital Authority of National Health Commission of the People’s Republic of China; Chinese Society of Oncology, Chinese Medical Association. Chinese protocol of diagnosis and treatment of colorectal cancer (2020 edition)[J]. Chin J Surg, 2020, 58(8): 601-625. | |
| [11] |
AGOSTINI F, ZAGALAK J, ATTIG J, et al. Intergenic RNA mainly derives from nascent transcripts of known genes[J]. Genome Biol, 2021, 22(1): 136.
doi: 10.1186/s13059-021-02350-x pmid: 33952325 |
| [12] |
ZHAO S, GUAN B, MI Y, et al. LncRNA MIR17HG promotes colorectal cancer liver metastasis by mediating a glycolysis-associated positive feedback circuit[J]. Oncogene, 2021, 40(28): 4709-4724.
doi: 10.1038/s41388-021-01859-6 pmid: 34145399 |
| [13] |
ZHU Y, HU H, YUAN Z, et al. LncRNA NEAT1 remodels chromatin to promote the 5-FU resistance by maintaining colorectal cancer stemness[J]. Cell Death Dis, 2020, 11(11): 962.
doi: 10.1038/s41419-020-03164-8 pmid: 33168814 |
| [14] |
LI Z J, CHENG J, SONG Y, et al. LncRNA SNHG5 upregulation induced by YY1 contributes to angiogenesis via miR-26b/CTGF/VEGFA axis in acute myelogenous leukemia[J]. Lab Invest, 2021, 101(3): 341-352.
doi: 10.1038/s41374-020-00519-9 |
| [15] |
GUO Q, DONG L, ZHANG C, et al. MicroRNA-363-3p, negatively regulated by long non-coding RNA small nucleolar RNA host gene 5, inhibits tumor progression by targeting Aurora kinase A in colorectal cancer[J]. Bioengineered, 2022, 13(3): 5357-5372.
doi: 10.1080/21655979.2021.2018972 pmid: 35166647 |
| [16] |
WANG Z, LIU T, XUE W, et al. ARNTL2 promotes pancreatic ductal adenocarcinoma progression through TGF/BETA pathway and is regulated by miR-26a-5p[J]. Cell Death Dis, 2020, 11(8): 692.
doi: 10.1038/s41419-020-02839-6 pmid: 32826856 |
| [17] | KONG W Q, LIANG J J, DU J, et al. Long noncoding RNA DLX6-AS1 regulates the growth and aggressiveness of colorectal cancer cells via mediating the miR-26a/EZH2 axis[J]. Cancer Biother Radiopharm, 2021, 36(9): 753-764. |
| [18] |
YANG Y, XI L, MA Y, et al. The lncRNA small nucleolar RNA host gene 5 regulates trophoblast cell proliferation, invasion, and migration via modulating miR-26a-5p/N-cadherin axis[J]. J Cell Biochem, 2019, 120(3): 3173-3184.
doi: 10.1002/jcb.27583 pmid: 30242892 |
| [19] | LI P P, LI R G, HUANG Y Q, et al. LncRNA OTUD6B-AS1 promotes paclitaxel resistance in triple negative breast cancer by regulation of miR-26a-5p/MTDH pathway-mediated autophagy and genomic instability[J]. Aging (Albany NY), 2021, 13(21): 24171-24191. |
| [20] |
KHAN M, SARKAR D. The scope of astrocyte elevated gene-1/metadherin (AEG-1/MTDH) in cancer clinicopathology: a review[J]. Genes (Basel), 2021, 12(2): 308.
doi: 10.3390/genes12020308 |
| [21] |
CHEN Y Y, HUANG S, GUO R, et al. Metadherin-mediated mechanisms in human malignancies[J]. Biomark Med, 2021, 15(18): 1769-1783.
doi: 10.2217/bmm-2021-0298 pmid: 34783585 |
| [22] | KLINGLER J, ANTON H, RÉAL E, et al. How HIV-1 gag manipulates its host cell proteins: a focus on interactors of the nucleocapsid domain[J]. Viruses, 2020, 12(8): E888. |
| [23] |
SETLAI B P, HULL R, REIS R M, et al. MicroRNA interrelated epithelial mesenchymal transition (EMT) in glioblastoma[J]. Genes (Basel), 2022, 13(2): 244.
doi: 10.3390/genes13020244 |
| [24] | HE A B, HE S M, HUANG C, et al. MTDH promotes metastasis of clear cell renal cell carcinoma by activating SND1-mediated ERK signaling and epithelial-mesenchymal transition[J]. Aging (Albany NY), 2020, 12(2): 1465-1487. |
| [25] |
ZHANG H, ZOU C, QIU Z, et al. CPEB3-mediated MTDH mRNA translational suppression restrains hepatocellular carcinoma progression[J]. Cell Death Dis, 2020, 11(9): 792.
doi: 10.1038/s41419-020-02984-y pmid: 32968053 |
| [1] | CHEN Yijun, LIU Yuhang, DUAN Haibo, WANG Xiongjun. Functional and mechanistic of AGPAT5 in liver cancer [J]. China Oncology, 2024, 34(9): 838-847. |
| [2] | HUANG Haozhe, CHEN Hong, ZHENG Dezhong, CHEN Chao, WANG Ying, XU Lichao, WANG Yaohui, HE Xinhong, YANG Yuanyuan, LI Wentao. A CT-based radiomics nomogram for predicting local tumor progression of colorectal cancer lung metastases treated with radiofrequency ablation [J]. China Oncology, 2024, 34(9): 857-872. |
| [3] | WU Wen, ZHANG Ruoxin, WENG Junyong, MA Yanlei, CAI Guoxiang, LI Xinxiang, YANG Yongzhi. Exploring the prognostic value of positive lymph node ratio in stage Ⅲ colorectal cancer patients and establishing a predictive model [J]. China Oncology, 2024, 34(9): 873-880. |
| [4] | GE Zuyin, SONG Kun, LIN Yunxiao, ZHONG Yeling, HAO Jingduo. The feasibility study of FCGBP and BIGH3 in circulating tumor cells as potential markers for colorectal cancer [J]. China Oncology, 2024, 34(8): 745-752. |
| [5] | CAO Xiaoshan, YANG Beibei, CONG Binbin, LIU Hong. The progress of treatment for brain metastases of triple-negative breast cancer [J]. China Oncology, 2024, 34(8): 777-784. |
| [6] | HUANG Sijie, KANG Xun, LI Wenbin. Clinical research progress of intrathecal therapy in the treatment of leptomeningeal metastasis [J]. China Oncology, 2024, 34(7): 695-701. |
| [7] | WENG Junyong, YE Zilan, ZHANG Ruoxin, LIU Qi, LI Xinxiang. Exploring the guiding role of the number of adverse pathological features in risk stratification for recurrence of stage Ⅰ-Ⅲ colorectal cancer: a retrospective cohort study of 9 875 cases [J]. China Oncology, 2024, 34(6): 527-536. |
| [8] | ZHANG Ruoxin, YE Zilan, WENG Junyong, LI Xinxiang. Correlation study between advanced age and inferior prognosis in stage Ⅱ colorectal cancer patients [J]. China Oncology, 2024, 34(5): 485-492. |
| [9] | SHEN Jie, FENG Xiaoshuang, WEN Hao, ZHOU Changming, MO Miao, WANG Zezhou, YUAN Jing, WU Xiaohua, ZHENG Ying. Metastasis patterns and survival analysis of 572 patients with metastatic cervical cancer: a hospital-based real world study [J]. China Oncology, 2024, 34(4): 361-367. |
| [10] | LI Xiaohui, ZHAO Jiaxu, PENG Haibao, ZHANG Ye, ZENG Rui, CHI Yudan. Effects of HMGA2 on migration and proliferation of leptomeningeal metastatic melanoma [J]. China Oncology, 2024, 34(4): 389-399. |
| [11] | LU Yue, LU Renquan, ZHANG Jie, ZHENG Hui. Application value of combined coagulation function indicators in monitoring hypercoagulable state of patients with colorectal cancer after chemotherapy [J]. China Oncology, 2024, 34(3): 278-285. |
| [12] | LI Jun, LU Tingwei, FANG Xuqian. Impact of MSI-H/dMMR on clinicopathological characteristics and prognosis of patients with BRAF V600E-mutated resectable colorectal cancer [J]. China Oncology, 2024, 34(11): 1061-1066. |
| [13] | WU Zhibai, XU Guiqin, ZHANG Li, YANG Zhaojuan, LIU Yun, JIAO Kun, CHEN Zehong, XU Chen, ZUO You, ZHENG Ningqian, YE Zhiqian, LIU Yongzhong. Mechanism study of KCMF1 promoting proliferation and NF-κB signaling transduction in colorectal cancer cells [J]. China Oncology, 2024, 34(11): 987-997. |
| [14] | OUYANG Fei, WANG Yang, CHEN Yu, PEI Guoqing, WANG Ling, ZHANG Yang, SHI Lei. Construction of the prediction model of breast cancer bone metastasis based on machine learning [J]. China Oncology, 2024, 34(10): 903-914. |
| [15] | ZHAO Junxiu, ZHU Yi, SONG Xiaoyu, ZHE Chao, XIAO Yuhan, LIU Yunduo, LI Linhai, XIAO Bin. Circ-0007766 acts as a miR-1972 sponge to promote breast cancer cell migration and invasion via upregulation of HER2 [J]. China Oncology, 2024, 34(10): 915-930. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
沪ICP备12009617
Powered by Beijing Magtech Co. Ltd
