Welcome to China Oncology,
China Oncology ›› 2024, Vol. 34 ›› Issue (6): 548-560.doi: 10.19401/j.cnki.1007-3639.2024.06.003
• Article • Previous Articles Next Articles
TANG Nan1( ), HUANG Huixia2,3,4(
), HUANG Huixia2,3,4( ), LIU Xiaojian1(
), LIU Xiaojian1( )(
)( )
)
Received:2024-02-06
Revised:2024-06-05
Online:2024-06-30
Published:2024-07-16
Share article
CLC Number:
TANG Nan, HUANG Huixia, LIU Xiaojian. Integrated single-cell sequencing and transcriptome sequencing to reveal a 9-gene prognostic signature of immune cells in colorectal cancer[J]. China Oncology, 2024, 34(6): 548-560.
Tab. 1
The primer list used in RTFQ-PCR"
| Primer | Sequence (5‘-3’) |
|---|---|
| CD36-1-F | GGCTGTGACCGGAACTGTG |
| CD36-1-R | AGGTCTCCAACTGGCATTAGAA |
| CTSD-F | ATTCAGGGCGAGTACATGATCC |
| CTSD-R | CGACACCTTGAGCGTGTAG |
| G0S2-F | TCGGCCTGATGGAGACTGT |
| G0S2-R | TTCTGGAGAGCCTGTCGCT |
| TIMP1-F | AGAGTGTCTGCGGATACTTCC |
| TIMP1-R | CCAACAGTGTAGGTCTTGGTG |
| HSPA1A-F | AGCTGGAGCAGGTGTGTAAC |
| HSPA1A-R | TACCTCCTCAATGGTGGGGC |
| VSIG4-1-F | GGGGCACCTAACAGTGGAC |
| VSIG4-1-R | GTCTGAGCCACGTTGTACCAG |
| CLEC4A-F | GACTGTGCTAGAATGGAGGCT |
| CLEC4A-R | GTCGCTGACCTTCTGGATCTG |
| TKT-1-F | TCATCGAGTGCTACATTGCTG |
| TKT-1-R | GCCATGCGAATCTGGTCAAAG |
| ASAH1-1-F | AGATGTCATGTGGATAGGGTTCC |
| ASAH1-1-R | GGGGCCAATATCTTGGTCTTG |
| β-actin-F | TTGTTACAGGAAGTCCCTTGCC |
| β-actin-R | ATGCTATCACCTCCCCTGTGTG |

Fig. 4
Measurement of signature gene expression in CRC tissues and cells A: RNA expression of TKT, TIMP1, G0S2, HSPA1A, CLEC4A, CD36, CTSD, VSIG4 and ASAH1 in tumor and paracancerous tissues. B: RNA expression of TKT, TIMP1, G0S2, HSPA1A, CLEC4A, CD36, CTSD, VSIG4 and ASAH1 in the mixed CRC cell lines (HCT116, HCT15, CACO2, HCT8, SW620, SW480, SW1116, RKO and DLD1) and the normal colon cell line NCM460. C: Representative images of immunohistochemical staining of TKT, TIMP1 and G0S2 in tumor and paracancerous tissues (×20, DAB; ×40) *: P<0.05, compared with each other; **: P<0.01, compared with each other; ***: P<0.001, compared with each other; ****: P<0.0001, compared with each other."
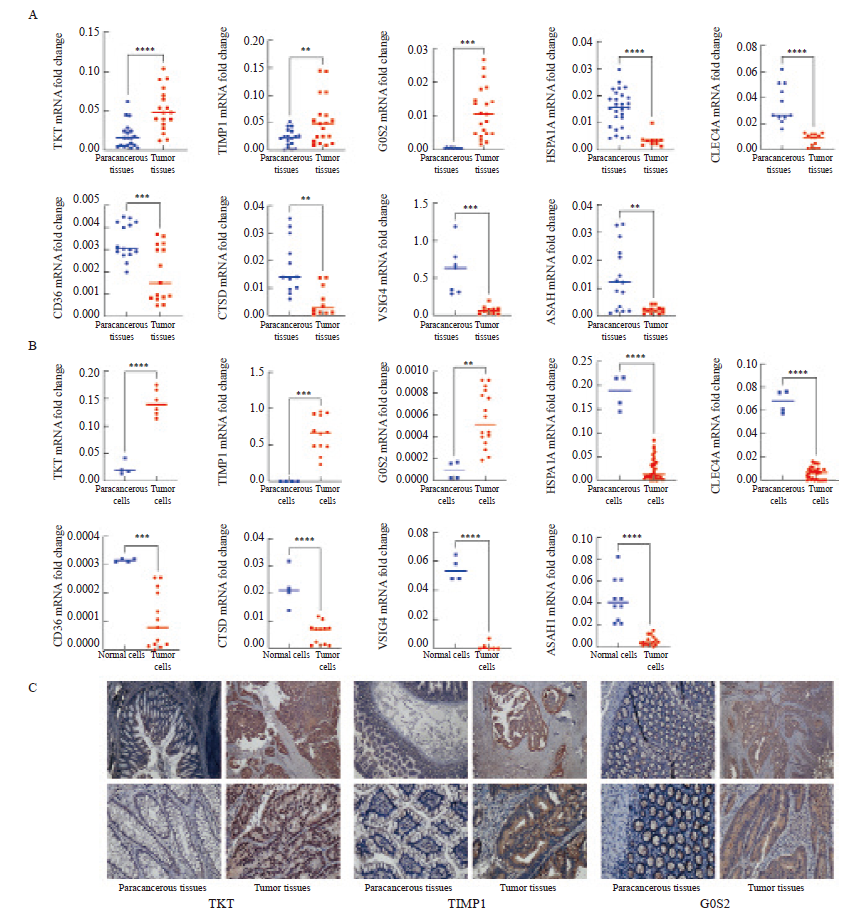

Fig. 5
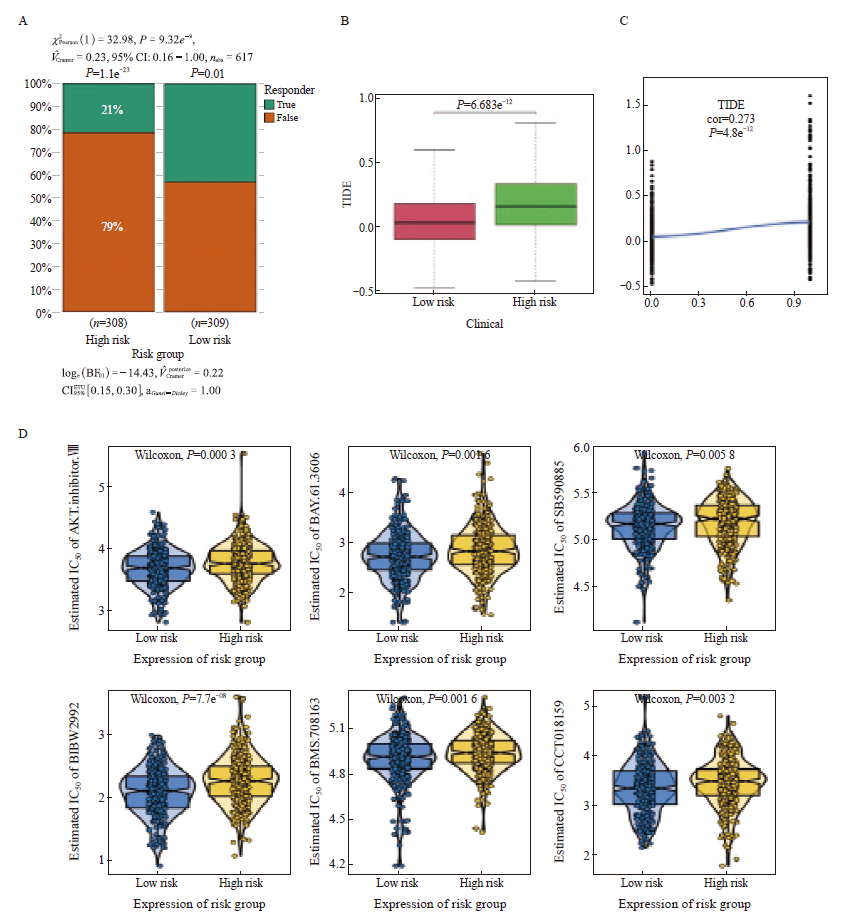
Immunotherapy response and chemotherapy sensitivity prediction A: Histogram of the response to immunotherapy in the high- and low-risk groups. B: Boxplot of TIDE scores in the high- and low-risk groups. C: Scatterplot of correlations between TIDE scores and risk scores. D: The IC50 values of 6 anti-cancer drugs of 2 risk groups of CRC patients."
| [1] |
DEKKER E, TANIS P J, VLEUGELS J L A, et al. Colorectal cancer[J]. Lancet, 2019, 394(10207): 1467-1480.
doi: S0140-6736(19)32319-0 pmid: 31631858 |
| [2] |
EDWARDS B K, NOONE A M, MARIOTTO A B, et al. Annual Report to the Nation on the status of cancer, 1975-2010, featuring prevalence of comorbidity and impact on survival among persons with lung, colorectal, breast, or prostate cancer[J]. Cancer, 2014, 120(9): 1290-1314.
doi: 10.1002/cncr.28509 pmid: 24343171 |
| [3] | PATEL S G, KARLITZ J J, YEN T, et al. The rising tide of early-onset colorectal cancer: a comprehensive review of epidemiology, clinical features, biology, risk factors, prevention, and early detection[J]. Lancet Gastroenterol Hepatol, 2022, 7(3): 262-274. |
| [4] |
HAN L, WANG S Y, WEI C, et al. Tumour microenvironment: a non-negligible driver for epithelial-mesenchymal transition in colorectal cancer[J]. Expert Rev Mol Med, 2021, 23: e16.
doi: 10.1017/erm.2021.13 pmid: 34758892 |
| [5] | LUO X J, ZHAO Q, LIU J, et al. Novel genetic and epigenetic biomarkers of prognostic and predictive significance in stage Ⅱ/Ⅲ colorectal cancer[J]. Mol Ther, 2021, 29(2): 587-596. |
| [6] |
CHEN B, SCURRAH C R, MCKINLEY E T, et al. Differential pre-malignant programs and microenvironment chart distinct paths to malignancy in human colorectal polyps[J]. Cell, 2021, 184(26): 6262-6280.e26.
doi: 10.1016/j.cell.2021.11.031 pmid: 34910928 |
| [7] | GUO W, ZHANG C Y, WANG X, et al. Resolving the difference between left-sided and right-sided colorectal cancer by single-cell sequencing[J]. JCI Insight, 2022, 7(1): e152616. |
| [8] | NORKIN M, ORDÓÑEZ-MORÁN P, HUELSKEN J. High-content, targeted RNA-seq screening in organoids for drug discovery in colorectal cancer[J]. Cell Rep, 2021, 35(3): 109026. |
| [9] | SIEGEL R L, MILLER K D, WAGLE N S, et al. Cancer statistics, 2023[J]. CA Cancer J Clin, 2023, 73(1): 17-48. |
| [10] | JIN M Z, JIN W L. The updated landscape of tumor microenvironment and drug repurposing[J]. Sig Transduct Target Ther, 2020, 5: 166. |
| [11] |
PITT J M, MARABELLE A, EGGERMONT A, et al. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy[J]. Ann Oncol, 2016, 27(8): 1482-1492.
doi: 10.1093/annonc/mdw168 pmid: 27069014 |
| [12] |
JACKSTADT R, VAN HOOFF S R, LEACH J D, et al. Epithelial NOTCH signaling rewires the tumor microenvironment of colorectal cancer to drive poor-prognosis subtypes and metastasis[J]. Cancer Cell, 2019, 36(3): 319-336.e7.
doi: S1535-6108(19)30371-X pmid: 31526760 |
| [13] | TIAN S B, CHU Y N, HU J, et al. Tumour-associated neutrophils secrete AGR2 to promote colorectal cancer metastasis via its receptor CD98hc-xCT[J]. Gut, 2022, 71(12): 2489-2501. |
| [14] | YIN Y, LIU B X, CAO Y L, et al. Colorectal cancer-derived small extracellular vesicles promote tumor immune evasion by upregulating PD-L1 expression in tumor-associated macrophages (adv. sci. 9/2022)[J]. Adv Sci, 2022, 9(9). |
| [15] | DEL VECCHIO F, MASTROIACO V, MARCO A D, et al. Next-generation sequencing: recent applications to the analysis of colorectal cancer[J]. J Transl Med, 2017, 15(1): 246. |
| [16] | LIANG L L, YU J, LI J, et al. Integration of scRNA-seq and bulk RNA-seq to analyse the heterogeneity of ovarian cancer immune cells and establish a molecular risk model[J]. Front Oncol, 2021, 11: 711020. |
| [17] | QU X D, ZHAO X Y, LIN K X, et al. M2-like tumor-associated macrophage-related biomarkers to construct a novel prognostic signature, reveal the immune landscape, and screen drugs in hepatocellular carcinoma[J]. Front Immunol, 2022, 13: 994019. |
| [18] | LU J, CHEN Y F, ZHANG X Q, et al. A novel prognostic model based on single-cell RNA sequencing data for hepatocellular carcinoma[J]. Cancer Cell Int, 2022, 22(1): 38. |
| [19] | ZHENG X B, SONG J N, YU C E, et al. Single-cell transcriptomic profiling unravels the adenoma-initiation role of protein tyrosine kinases during colorectal tumorigenesis[J]. Signal Transduct Target Ther, 2022, 7(1): 60. |
| [20] |
VIJAYAN Y, LANKADASARI M B, HARIKUMAR K B. Acid ceramidase: a novel therapeutic target in cancer[J]. Curr Top Med Chem, 2019, 19(17): 1512-1520.
doi: 10.2174/1568026619666190227222930 pmid: 30827244 |
| [21] |
LUCKI N C, SEWER M B. Genistein stimulates MCF-7 breast cancer cell growth by inducing acid ceramidase (ASAH1) gene expression[J]. J Biol Chem, 2011, 286(22): 19399-19409.
doi: 10.1074/jbc.M110.195826 pmid: 21493710 |
| [22] | LI Y H, LIU H T, XU J, et al. The value of detection of S100A8 and ASAH1 in predicting the chemotherapy response for breast cancer patients[J]. Hum Pathol, 2018, 74: 156-163. |
| [23] |
REALINI N, SOLORZANO C, PAGLIUCA C, et al. Discovery of highly potent acid ceramidase inhibitors with in vitro tumor chemosensitizing activity[J]. Sci Rep, 2013, 3: 1035.
doi: 10.1038/srep01035 pmid: 23301156 |
| [24] | MACHALA M, PROCHÁZKOVÁ J, HOFMANOVÁ J, et al. Colon cancer and perturbations of the sphingolipid metabolism[J]. Int J Mol Sci, 2019, 20(23): 6051. |
| [25] | VIJAYAN Y, JAMES S, VISWANATHAN A, et al. Targeting acid ceramidase enhances antitumor immune response in colorectal cancer[J]. J Adv Res, 2023: S 2090-S1232(23)00403-4. |
| [26] | LI M L, ZHAO X, YONG H M, et al. Transketolase promotes colorectal cancer metastasis through regulating AKT phosphorylation[J]. Cell Death Dis, 2022, 13(2): 99. |
| [27] |
YANG H, WU X L, WU K H, et al. MicroRNA-497 regulates cisplatin chemosensitivity of cervical cancer by targeting transketolase[J]. Am J Cancer Res, 2016, 6(11): 2690-2699.
pmid: 27904781 |
| [28] | SHUKLA S K, PUROHIT V, MEHLA K, et al. MUC1 and HIF-1alpha signaling crosstalk induces anabolic glucose metabolism to impart gemcitabine resistance to pancreatic cancer[J]. Cancer Cell, 2017, 32(3): 392. |
| [29] | DASGUPTA S, RAJAPAKSHE K, ZHU B K, et al. Metabolic enzyme PFKFB4 activates transcriptional coactivator SRC-3 to drive breast cancer[J]. Nature, 2018, 556(7700): 249-254. |
| [30] | WANG H L, CHEN Y, WANG Y Q, et al. Sirtuin5 protects colorectal cancer from DNA damage by keeping nucleotide availability[J]. Nat Commun, 2022, 13(1): 6121. |
| [31] | UTO T, FUKAYA T, MITOMA S, et al. Clec4A4 acts as a negative immune checkpoint regulator to suppress antitumor immunity[J]. Cancer Immunol Res, 2023, 11(9): 1266-1279. |
| [32] | LIU B, CHENG L, GAO H H, et al. The biology of VSIG4: implications for the treatment of immune-mediated inflammatory diseases and cancer[J]. Cancer Lett, 2023, 553: 215996. |
| [33] | CHOW A, SCHAD S, GREEN M D, et al. Tim-4+ cavity-resident macrophages impair anti-tumor CD8+ T cell immunity[J]. Cancer Cell, 2021, 39(7): 973-988.e9. |
| [34] | WU S, PEI Q, NI W, et al. HSPA1A protects cells from thermal stress by impeding ESCRT-0-mediated autophagic flux in epidermal thermoresistance[J]. J Investig Dermatol, 2021, 141(1): 48-58.e3. |
| [35] | WANG X, WANG Y T, FANG Z Y, et al. Targeting HSPA1A in ARID2-deficient lung adenocarcinoma[J]. Natl Sci Rev, 2021, 8(10): nwab014. |
| [36] | JIANG Y, XU Y J, ZHENG C, et al. Acetyltransferase from Akkermansia muciniphilablunts colorectal tumourigenesis by reprogramming tumour microenvironment[J]. Gut, 2023, 72(7): 1308-1318. |
| [37] |
GUCCINI I, REVANDKAR A, D’AMBROSIO M, et al. Senescence reprogramming by TIMP1 deficiency promotes prostate cancer metastasis[J]. Cancer Cell, 2021, 39(1): 68-82.e9.
doi: 10.1016/j.ccell.2020.10.012 pmid: 33186519 |
| [38] | TIAN Z F, OU G S, SU M X, et al. TIMP1 derived from pancreatic cancer cells stimulates Schwann cells and promotes the occurrence of perineural invasion[J]. Cancer Lett, 2022, 546: 215863. |
| [39] | SCHOEPS B, ECKFELD C, PROKOPCHUK O, et al. TIMP1 triggers neutrophil extracellular trap formation in pancreatic cancer[J]. Cancer Res, 2021, 81(13): 3568-3579. |
| [40] | MA B B, UEDA H, OKAMOTO K, et al. TIMP1 promotes cell proliferation and invasion capability of right-sided colon cancers via the FAK/Akt signaling pathway[J]. Cancer Sci, 2022, 113(12): 4244-4257. |
| [41] | HECKMANN B L, ZHANG X D, XIE X T, et al. The G0/G1 switch gene 2 (G0S2): regulating metabolism and beyond[J]. Biochim Biophys Acta, 2013, 1831(2): 276-281. |
| [42] |
NIELSEN T S, MØLLER N. Adipose triglyceride lipase and G0/G1 switch gene 2: approaching proof of concept[J]. Diabetes, 2014, 63(3): 847-849.
doi: 10.2337/db13-1838 pmid: 24556865 |
| [43] | KIOKA H, KATO H, FUJIKAWA M, et al. Evaluation of intramitochondrial ATP levels identifies G0/G1 switch gene 2 as a positive regulator of oxidative phosphorylation[J]. Proc Natl Acad Sci U S A, 2014, 111(1): 273-278. |
| [44] |
CHANG X F, MONITTO C L, DEMOKAN S, et al. Identification of hypermethylated genes associated with cisplatin resistance in human cancers[J]. Cancer Res, 2010, 70(7): 2870-2879.
doi: 10.1158/0008-5472.CAN-09-3427 pmid: 20215521 |
| [45] | BARREAU O, ASSIÉ G, WILMOT-ROUSSEL H, et al. Identification of a CpG island methylator phenotype in adrenocortical carcinomas[J]. J Clin Endocrinol Metab, 2013, 98(1): E174-E184. |
| [46] |
KUSAKABE M, KUTOMI T, WATANABE K, et al. Identification of G0S2 as a gene frequently methylated in squamous lung cancer by combination of in silico and experimental approaches[J]. Int J Cancer, 2010, 126(8): 1895-1902.
doi: 10.1002/ijc.24947 pmid: 19816938 |
| [47] | ASHRAF Y, MANSOURI H, LAURENT-MATHA V, et al. Immunotherapy of triple-negative breast cancer with cathepsin D-targeting antibodies[J]. J Immunother Cancer, 2019, 7(1): 29. |
| [48] | LIU C M, SHEN H T, LIN Y A, et al. Antiproliferative and antimetastatic effects of praeruptorin C on human non-small cell lung cancer through inactivating ERK/CTSD signalling pathways[J]. Molecules, 2020, 25(7): 1625. |
| [49] | XU J S, DAI S Q, YUAN Y, et al. A prognostic model for colon cancer patients based on eight signature autophagy genes[J]. Front Cell Dev Biol, 2020, 8: 602174. |
| [50] | XIE L Q, ZHAO C, CAI S J, et al. Novel proteomic strategy reveal combined alpha1 antitrypsin and cathepsin D as biomarkers for colorectal cancer early screening[J]. J Proteome Res, 2010, 9(9): 4701-4709. |
| [51] | WANG K, FU S Y, DONG L X, et al. Periplocin suppresses the growth of colorectal cancer cells by triggering LGALS3 (galectin 3)-mediated lysophagy[J]. Autophagy, 2023, 19(12): 3132-3150. |
| [1] | WANG Zifei, DING Yahui, LI Yan, LUAN Xin, TANG Min. Application of 3D bioprinting in cancer research and tissue engineering [J]. China Oncology, 2024, 34(9): 814-826. |
| [2] | CHEN Hong, CAO Zhiyun. Recent progress in the construction and application of patient-derived pancreatic cancer organoid models [J]. China Oncology, 2024, 34(6): 590-597. |
| [3] | JIANG Yuanyuan, WEI Wenfei, WU Jingya, LI Huawen. Application of organoids in drug screening of gynecological malignant tumors [J]. China Oncology, 2024, 34(11): 1053-1060. |
| [4] | WANG Ruotong, WANG Xin, SHEN Bo. Application and progress of organoids in tumor translational medicine [J]. China Oncology, 2022, 32(11): 1105-1114. |
| [5] | SHANG Kun, ZHANG Peng, ZHOU Lu, WANG Zhen, CAO Yue, LI Ying-yi. Screening of Pim kinase inhibitors by the establishment of high-throughput ELISA system [J]. China Oncology, 2013, 23(4): 260-266. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
沪ICP备12009617
Powered by Beijing Magtech Co. Ltd
