Welcome to China Oncology,
China Oncology ›› 2022, Vol. 32 ›› Issue (12): 1218-1228.doi: 10.19401/j.cnki.1007-3639.2022.12.010
• Article • Previous Articles Next Articles
LIAO Xinghe1( ), LIU Zhantao2, LIU Minghui1
), LIU Zhantao2, LIU Minghui1
Received:2022-09-25
Revised:2022-12-01
Online:2022-12-30
Published:2023-02-02
Contact:
LIAO Xinghe
Share article
CLC Number:
LIAO Xinghe, LIU Zhantao, LIU Minghui. Glioma stem cell-derived exosomal lncRNA HOXA-AS2 promoted proliferation, migration, invasion and stemness in glioma[J]. China Oncology, 2022, 32(12): 1218-1228.
Fig. 1
Identification of DElncRNAs in glioma A, B: Volcano plot of DelncRNAs in CGGA and TCGA; C: 11 overlapping DelncRNAs were identified using R language; D: The correlation between HOXA-AS2 level and OS rate in patients with glioma in the CGGA datasets; E: The correlation between HOXA-AS2 level and OS rate in patients with glioma in the TCGA datasets; F: RTFQ-PCR showing HOXA-AS2 levels in T98G, SHG44 and U251 MG cells. **: P <0.01, compared with HA1800 group."
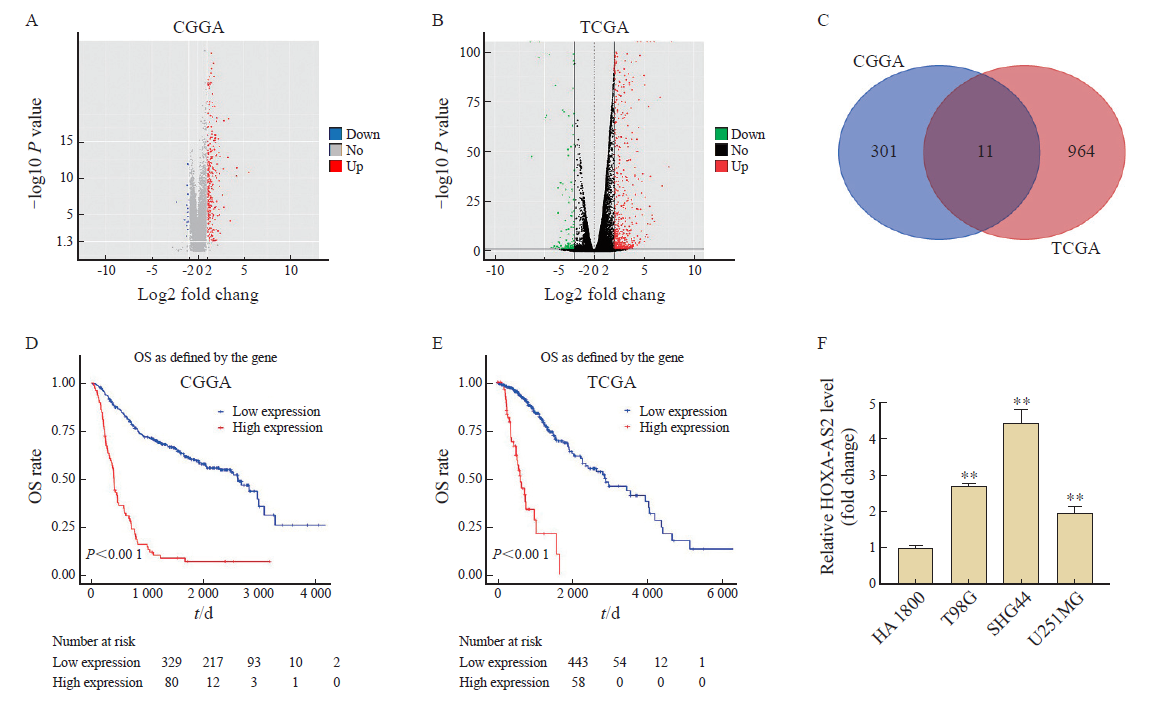
Fig. 2
HOXA-AS2 is upregulated in GSC A: Isolation of CD133+ cells (SHG44-GSC) from SHG44 cells using flow cytometry. Meanwhile, the percentage of CD133+ cells in the SHG44-GSC and SHG44 cells were detected; B: Western blot analysis of CD133, SOX2, OCT4 protein expressions in SHG44 cells and SHG44-GSC cells; C: RTFQ-PCR showing HOXA-AS2 levels in SHG44 cells and SHG44-GSC cells. **: P<0.01, compared with SHG44 cell group."
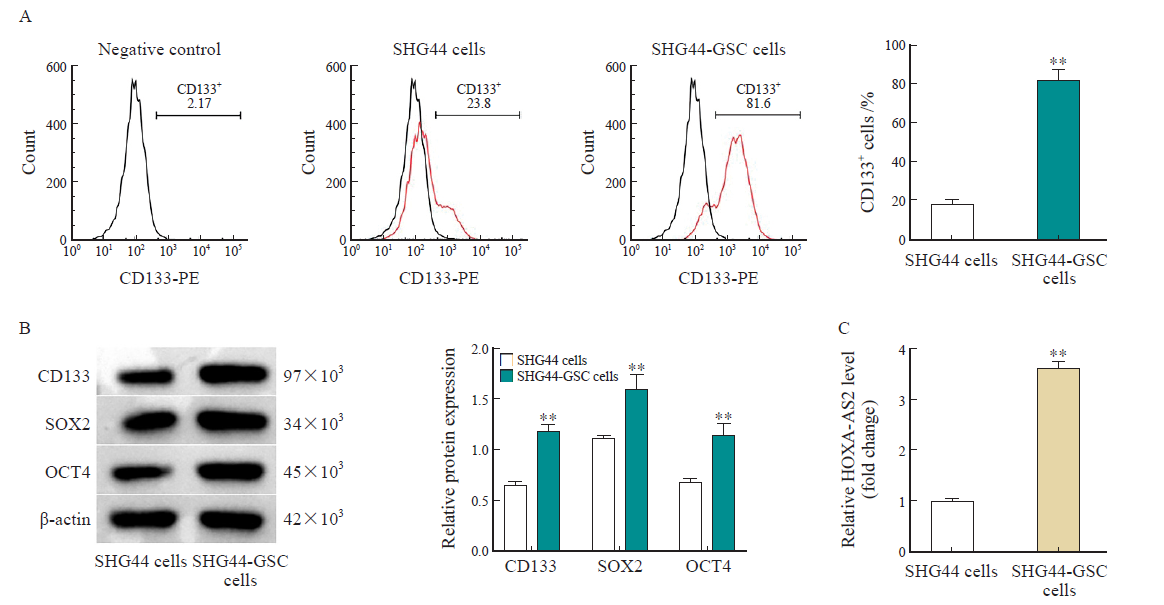
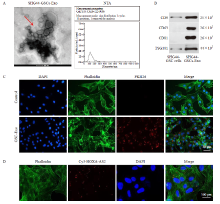
Fig. 3
Characterization of exosomal particles A: NTA and TEM assays were used to identify exosomes; B: Exosome surface markers (CD9, CD63, CD81 and TSG101) detected by Western blot analysis; C: The exosomes absorbed by SHG44 cells were observed under confocal fluorescence microscope. Red color: Exosome; Green color: SHG44 cells; Blue color: Cell nucleus. Control group: SHG44 cells without the treatment of exosomes; D: Cy3-labeled HOXA-AS2 in SHG44 cells were observed under confocal fluorescence microscope. Red color: Cy3-labeled HOXA-AS2; Green color: SHG44 cells; Blue color: Cell nucleus."
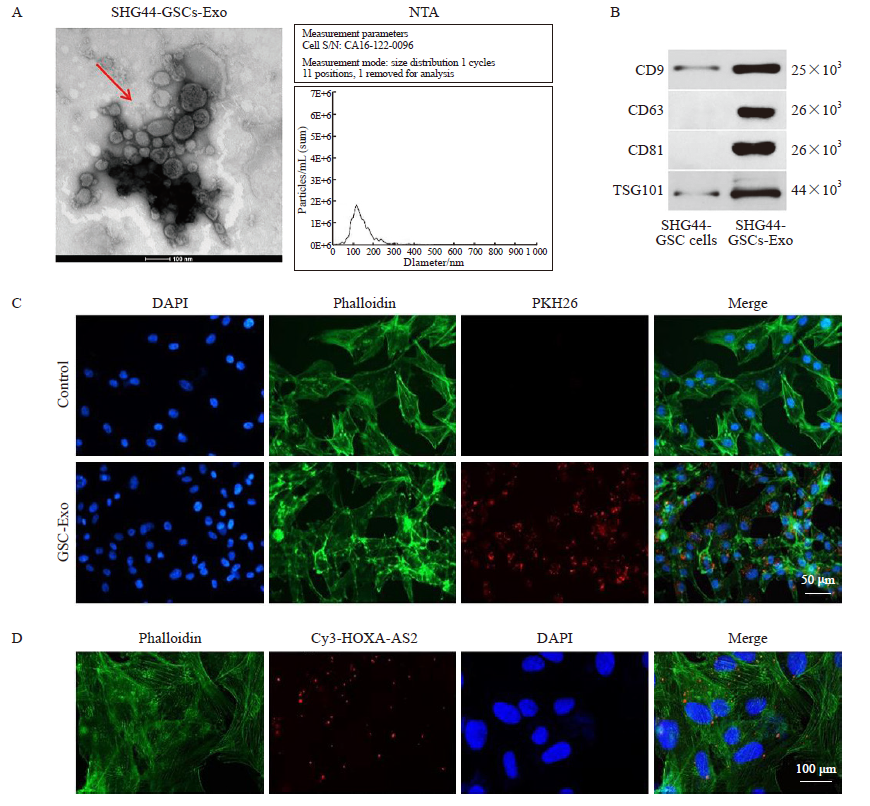
Fig. 4
SHG44-GSC transfer HOXA-AS2 to SHG44 cells by exosomes A: RTFQ-PCR showing HOXA-AS2 levels in SHG44-GSC transfected with HOXA-AS2 OE; B: RTFQ-PCR analysis of HOXA-AS2 level in SHG44-GSC transfected with HOXA-AS2 shRNA1 and HOXA-AS2 shRNA2; C: RTFQ-PCR analysis of HOXA-AS2 level in exosomes derived from SHG44-GSC that transfected with HOXA-AS2 shRNA1 and HOXA-AS2 shRNA2; D: RTFQ-PCR analysis of HOXA-AS2 level in SHG44 cells co-cultured with transfected SHG44-GSC; E: RTFQ-PCR showing HOXA-AS2 levels in SHG44 cells co-cultured with GW4869-treated transfected SHG44-GSC; F: RTFQ-PCR showing HOXA-AS2 levels in SHG44 cells treated with indicated exosomes. **: P<0.01, compared with each other."
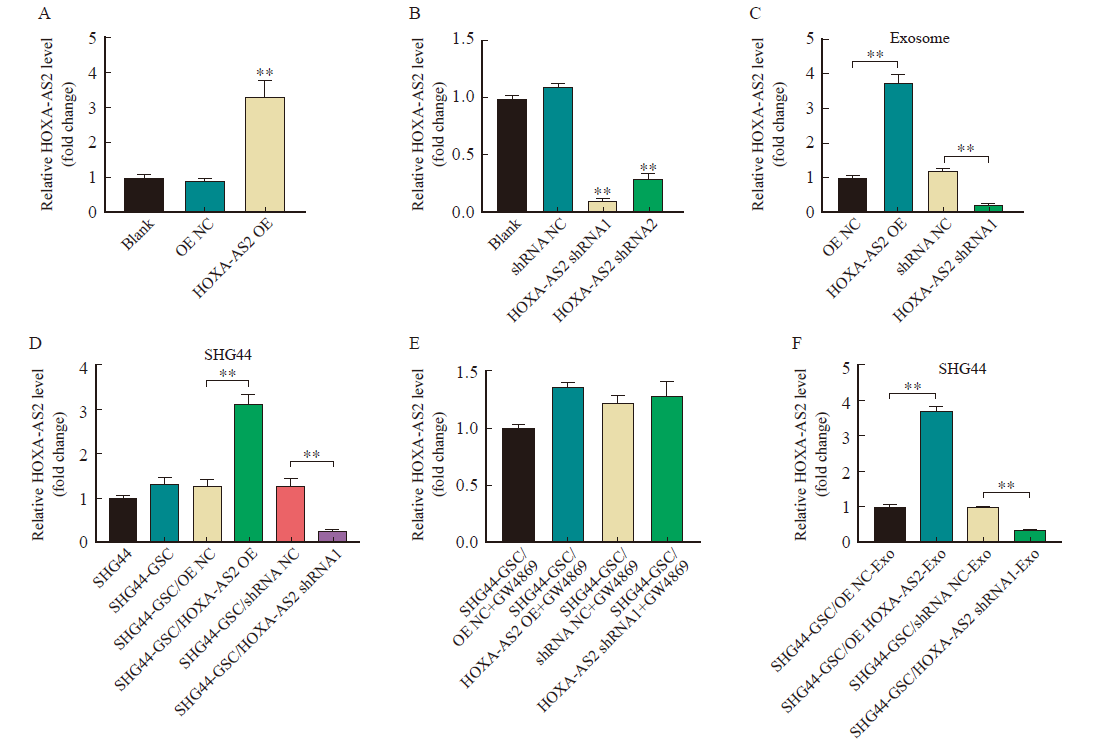
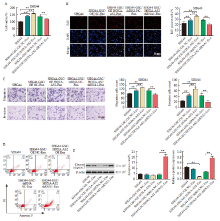
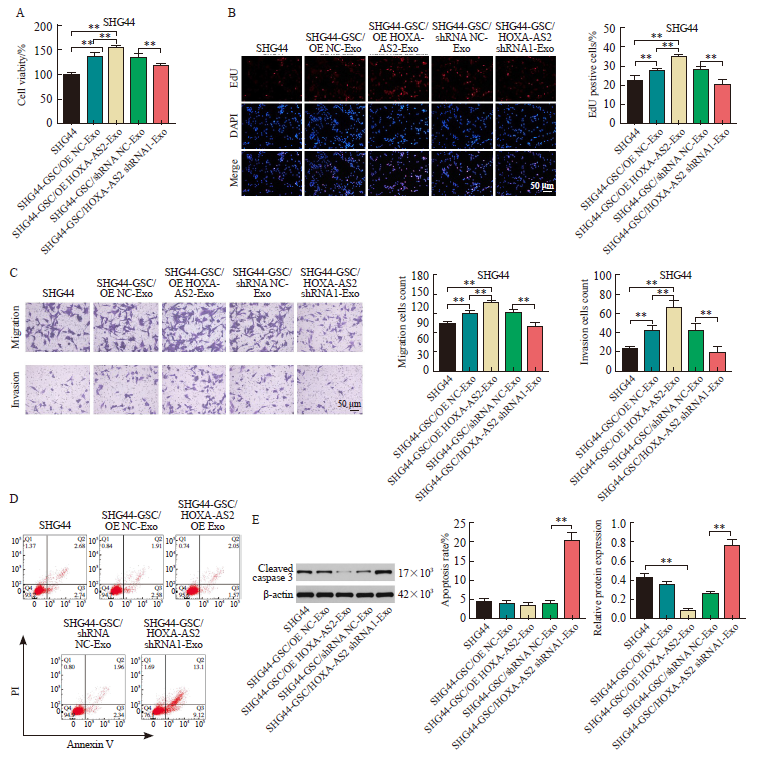
Fig. 5
HOXA-AS2 transferred by SHG44-GSC-derived exosomes promoted proliferation, migration, invasion of SHG44 cells A: CCK-8 assay of SHG44 cells incubated with indicated exosomes for 48 h; B: EdU staining assay of SHG44 cells incubated with indicated exosomes for 48 h; C: Transwell migration and invasion assays of SHG44 cells incubated with indicated exosomes for 24 h; D: Flow cytometry assay of SHG44 cells incubated with indicated exosomes for 48 h; E: Western blot showing cleaved caspase 3 expression in SHG44 cells incubated with indicated exosomes for 48 h. **: P<0.01, compared with each other."
| [1] |
OSTROM Q T, BAUCHET L, DAVIS F G, et al. The epidemiology of glioma in adults: a “state of the science” review[J]. Neuro Oncol, 2014, 16(7): 896-913.
doi: 10.1093/neuonc/nou087 |
| [2] |
MALTA T M, DE SOUZA C F, SABEDOT T S, et al. Glioma CpG island methylator phenotype (G-CIMP): biological and clinical implications[J]. Neuro Oncol, 2018, 20(5): 608-620.
doi: 10.1093/neuonc/nox183 |
| [3] |
SHI J, ZHANG Y, QIN B, et al. Long non-coding RNA LINC00174 promotes glycolysis and tumor progression by regulating miR-152-3p/SLC2A1 axis in glioma[J]. J Exp Clin Cancer Res, 2019, 38(1): 395.
doi: 10.1186/s13046-019-1390-x |
| [4] |
MIYAUCHI J T, TSIRKA S E. Advances in immunotherapeutic research for glioma therapy[J]. J Neurol, 2018, 265(4): 741-756.
doi: 10.1007/s00415-017-8695-5 pmid: 29209782 |
| [5] | LV Q L, WANG L C, LI D C, et al. Knockdown lncRNA DLEU1 inhibits gliomas progression and promotes temozolomide chemosensitivity by regulating autophagy[J]. Front Pharmacol, 2020, 11: 560543. |
| [6] |
FU C, LI D Y, ZHANG X N, et al. LncRNA PVT1 facilitates tumorigenesis and progression of glioma via regulation of miR-128-3p/GREM1 axis and BMP signaling pathway[J]. Neurotherapeutics, 2018, 15(4): 1139-1157.
doi: 10.1007/s13311-018-0649-9 pmid: 30120709 |
| [7] |
CHEN P Y, LI X D, MA W N, et al. Comprehensive transcriptomic analysis and experimental validation identify lncRNA HOXA-AS2/miR-184/COL6A2 as the critical ceRNA regulation involved in low-grade glioma recurrence[J]. Onco Targets Ther, 2020, 13: 4999-5016.
doi: 10.2147/OTT.S245896 |
| [8] | LIU X, ZHU Q J, GUO Y, et al. LncRNA LINC00689 promotes the growth, metastasis and glycolysis of glioma cells by targeting miR-338-3p/PKM2 axis[J]. Biomedecine Pharmacother, 2019, 117: 109069. |
| [9] |
AKELLA N M, LE MINH G, CIRAKU L, et al. O-GlcNAc transferase regulates cancer stem-like potential of breast cancer cells[J]. Mol Cancer Res, 2020, 18(4): 585-598.
doi: 10.1158/1541-7786.MCR-19-0732 pmid: 31974291 |
| [10] | MOHIUDDIN I S, WEI S J, KANG M H. Role of OCT4 in cancer stem-like cells and chemotherapy resistance[J]. Biochim Biophys Acta Mol Basis Dis, 2020, 1866(4): 165432. |
| [11] |
SUN Z, WANG L, DONG L H, et al. Emerging role of exosome signalling in maintaining cancer stem cell dynamic equilibrium[J]. J Cell Mol Med, 2018, 22(8): 3719-3728.
doi: 10.1111/jcmm.13676 |
| [12] |
LI W G, JIANG P F, SUN X L, et al. Suppressing H19 modulates tumorigenicity and stemness in U251 and U87MG glioma cells[J]. Cell Mol Neurobiol, 2016, 36(8): 1219-1227.
pmid: 26983719 |
| [13] | SASAKI R, KANDA T, YOKOSUKA O, et al. Exosomes and hepatocellular carcinoma: from bench to bedside[J]. Int J Mol Sci, 2019, 20(6): E1406. |
| [14] |
KAHLERT C, KALLURI R. Exosomes in tumor microenvironment influence cancer progression and metastasis[J]. J Mol Med (Berl), 2013, 91(4): 431-437.
doi: 10.1007/s00109-013-1020-6 pmid: 23519402 |
| [15] |
XIONG L, WANG F, XIE X Q. Advanced treatment in high-grade gliomas[J]. J BUON, 2019, 24(2): 424-430.
pmid: 31127986 |
| [16] | NAN Y. Molecular targeted therapy in glioblastoma[J]. J Int Oncol, 2017, 44: 689-692. |
| [17] |
KOPP F, MENDELL J T. Functional classification and experimental dissection of long noncoding RNAs[J]. Cell, 2018, 172(3): 393-407.
doi: S0092-8674(18)30048-5 pmid: 29373828 |
| [18] |
ANGELOPOULOU E, PAUDEL Y N, PIPERI C. Critical role of HOX transcript antisense intergenic RNA (HOTAIR) in gliomas[J]. J Mol Med (Berl), 2020, 98(11): 1525-1546.
doi: 10.1007/s00109-020-01984-x |
| [19] | 曲来强, 彭静, 喻磊, 等. lncRNA HOXA-AS2对视网膜母细胞瘤细胞增殖和上皮间质转化的影响[J]. 中国肿瘤临床与康复, 2021, 28(11): 1308-1311. |
| QU L Q, PENG J, YU L, et al. Effects of LncRNA HOXA-AS2 on the proliferation and epithelial-mesenchymal transition of retinoblastoma cells[J]. Chin J Clin Oncol Rehabilitation, 2021, 28(11): 1308-1311. | |
| [20] | 陈升阳, 陈艳军, 胡水全, 等. LncRNA HOXA-AS2在胰腺癌组织中的表达及对细胞增殖和侵袭力的影响[J]. 实用医学杂志, 2021, 37(14): 1795-1799. |
| CHEN S Y, CHEN Y J, HU S Q, et al. Expression of lncRNA HOXA-AS2 in pancreatic cancer tissues and its effect on cell proliferation and invasion[J]. J Pract Med, 2021, 37(14): 1795-1799. | |
| [21] |
ZHANG P, CAO P H, ZHU X F, et al. Upregulation of long non-coding RNA HOXA-AS2 promotes proliferation and induces epithelial-mesenchymal transition in gallbladder carcinoma[J]. Oncotarget, 2017, 8(20): 33137-33143.
doi: 10.18632/oncotarget.16561 pmid: 28388535 |
| [22] | 俞盛健, 麻怀露, 陈王洋, 等. 基于生物信息学的脑胶质瘤预后风险长链非编码RNA筛选[J]. 温州医科大学学报, 2020, 50(3): 199-205. |
| YU S J, MA H L, CHEN W Y, et al. Bioinformatics study on the screening of long-chain noncoding RNA for prognostic risk of glioma[J]. J Wenzhou Med Univ, 2020, 50(3): 199-205. | |
| [23] |
WU L X, ZHU X Q, SONG Z Y, et al. Long non-coding RNA HOXA-AS2 enhances the malignant biological behaviors in glioma by epigenetically regulating RND3 expression[J]. Onco Targets Ther, 2019, 12: 9407-9419.
doi: 10.2147/OTT.S225678 |
| [24] |
GAO Y N, YU H, LIU Y H, et al. Long non-coding RNA HOXA-AS2 regulates malignant glioma behaviors and vasculogenic mimicry formation via the miR-373/EGFR axis[J]. Cell Physiol Biochem, 2018, 45(1): 131-147.
doi: 10.1159/000486253 pmid: 29310118 |
| [25] | DAI J, SU Y Z, ZHONG S Y, et al. Exosomes: key players in cancer and potential therapeutic strategy[J]. Signal Transduct Target Ther, 2020, 5(1): 145. |
| [26] | XU J S, LIAO K L, ZHOU W M. Exosomes regulate the transformation of cancer cells in cancer stem cell homeostasis[J]. Stem Cells Int, 2018, 2018: 4837370. |
| [27] |
CONIGLIARO A, FONTANA S, RAIMONDO S, et al. Exosomes: nanocarriers of biological messages[J]. Adv Exp Med Biol, 2017, 998: 23-43.
doi: 10.1007/978-981-10-4397-0_2 pmid: 28936730 |
| [28] |
WANG S S, GAO X L, LIU X, et al. CD133+ cancer stem-like cells promote migration and invasion of salivary adenoid cystic carcinoma by inducing vasculogenic mimicry formation[J]. Oncotarget, 2016, 7(20): 29051-29062.
doi: 10.18632/oncotarget.8665 |
| [29] | CHEN W Q, ALLEN S G, QIAN W Y, et al. Biophysical phenotyping and modulation of ALDH + inflammatory breast cancer stem-like cells[J]. Small, 2019, 15(5): e1802891. |
| [30] |
JIANG Y, SONG Y F, WANG R, et al. NFAT1-mediated regulation of NDEL1 promotes growth and invasion of glioma stem-like cells[J]. Cancer Res, 2019, 79(10): 2593-2603.
doi: 10.1158/0008-5472.CAN-18-3297 pmid: 30940662 |
| [31] |
CHAI Y, WU H T, LIANG C D, et al. Exosomal lncRNA ROR1-AS1 derived from tumor cells promotes glioma progression via regulating miR-4686[J]. Int J Nanomedicine, 2020, 15: 8863-8872.
doi: 10.2147/IJN.S271795 |
| [32] |
YOUSEFI H, MAHERONNAGHSH M, MOLAEI F, et al. Long noncoding RNAs and exosomal lncRNAs: classification, and mechanisms in breast cancer metastasis and drug resistance[J]. Oncogene, 2020, 39(5): 953-974.
doi: 10.1038/s41388-019-1040-y pmid: 31601996 |
| [33] | JIANG J X, LU J, WANG X L, et al. Glioma stem cell-derived exosomal miR-944 reduces glioma growth and angiogenesis by inhibiting AKT/ERK signaling[J]. Aging (Albany NY), 2021, 13(15): 19243-19259. |
| [34] |
CONIGLIARO A, COSTA V, LO DICO A, et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA[J]. Mol Cancer, 2015, 14: 155.
doi: 10.1186/s12943-015-0426-x |
| [35] |
SUN Z, WANG L, ZHOU Y L, et al. Glioblastoma stem cell-derived exosomes enhance stemness and tumorigenicity of glioma cells by transferring Notch1 protein[J]. Cell Mol Neurobiol, 2020, 40(5): 767-784.
doi: 10.1007/s10571-019-00771-8 pmid: 31853695 |
| [1] | CAO Fei, YU Wenhao, TANG Xiaonan, MA Zidong, CHANG Tingmin, GONG Yabin, LIAO Mingjuan, KANG Xiaohong. Mechanism of LINC01410 promoting proliferation and migration in esophageal squamous cell carcinoma [J]. China Oncology, 2024, 34(8): 753-762. |
| [2] | YE Junling, ZHENG Xiaoying, GUO Xinjian, CHEN Ruihui, YANG Liu, GOU Xiaodan, JIANG Hanmei. A study on mechanism of lncRNA-mediated SNHG5/miR-26a-5p/MTDH signal axis promoting metastasis of colorectal cancer [J]. China Oncology, 2023, 33(7): 673-685. |
| [3] | HU Keshu, LIU Wenfeng, ZHANG Feng, QUAN Bing, YIN Xin. Mechanism of LINC00601 in regulating sensitivity of hepatocellular carcinoma cells to oxaliplatin chemotherapy [J]. China Oncology, 2023, 33(1): 25-35. |
| [4] | LÜ Ye, YING Yexia, YANG Wenjing, HOU Yujin, YUAN Chunxiu, CAO Xiangmei. The effect of DZNep on tumor microenvironment through regulating exosomes derived from invasive breast carcinoma [J]. China Oncology, 2022, 32(9): 827-835. |
| [5] | CHEN Anli, SHEN Haoyuan, YANG Shixiong, DENG Chunyan, HU Chaohua, LIU Hanzhong, WANG Shu, QIAN Fang. Predictive value of plasma long non-coding RNA H19 in neoadjuvant therapy for breast cancer [J]. China Oncology, 2022, 32(8): 727-735. |
| [6] | MU Jiaqian, TENG Xiaoyan, WEI Lirong, QIU Rong, GUI Pengcheng, DU Yuzhen. The role and application value of integrin β3 in bone metastasis of lung adenocarcinoma [J]. China Oncology, 2022, 32(4): 351-356. |
| [7] | JIN Youping, LI Luying, ZHOU Ping. A study on the role of long non-coding RNA STMN1P2 in breast cancer and its mechanism [J]. China Oncology, 2021, 31(7): 596-604. |
| [8] | PAN Xinghao, LIU Mingbin, ZHANG Xiaoyan, XU Jianqing. Advances in research of exosomes as targeted therapeutic vectors [J]. China Oncology, 2020, 30(9): 701-706. |
| [9] | ZHANG Lianglong , AN Hongwei , ZHAO Lizhi , DONG Qian , GAO Guiyan . Mechanism of overexpression of lncRNA LINC00152 in temozolomide-induced stem cell cycle arrest in glioma [J]. China Oncology, 2020, 30(5): 328-334. |
| [10] | HUANG Limin, LEI Zhu, ZHANG Yu, LI Yong, HOU Jing, XU Zhong, CAO Hui, WANG Tao, YANG Jie. Clinical evaluation of postoperative residual tumor by 3D-ASL magnetic resonance imaging in high-grade glioma [J]. China Oncology, 2019, 29(11): 875-879. |
| [11] | WANG Xiuyue, ZHAO Chuan, CHEN Che, et al. A meta-analysis of long non-coding RNAs as novel serum biomarkers for the diagnosis of hepatocellular carcinoma [J]. China Oncology, 2018, 28(3): 229-235. |
| [12] | CHI Hong, CHEN Junxia. The role of lncRNA RP11-79H23.3 in bladder cancer cells and its mechanism [J]. China Oncology, 2018, 28(1): 22-29. |
| [13] | LIU Lei,WU Jiong. Research progress of long non-coding RNA related to breast cancer [J]. China Oncology, 2017, 27(8): 668-674. |
| [14] | LIU Ning, CHENG Dongdong, JIANG Jinbo. The functional role of long non-coding RNA PANDAR in promoting colorectal cancer metastasis and its mechanism [J]. China Oncology, 2017, 27(4): 268-275. |
| [15] | HE Anbang, CHEN Haodong, Lü Zhaojie, et al. Research progress on long non-coding RNA in renal cell carcinoma [J]. China Oncology, 2016, 26(8): 704-711. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
沪ICP备12009617
Powered by Beijing Magtech Co. Ltd
