Welcome to China Oncology,
China Oncology ›› 2022, Vol. 32 ›› Issue (4): 298-308.doi: 10.19401/j.cnki.1007-3639.2022.04.002
• Article • Previous Articles Next Articles
CAI Juan1( )(
)( ), CHEN Zhiqiang2, ZUO Xueliang3(
), CHEN Zhiqiang2, ZUO Xueliang3( )(
)( )
)
Received:2021-11-20
Revised:2022-03-01
Online:2022-04-30
Published:2022-05-07
Contact:
ZUO Xueliang
E-mail:caijuan1987@yeah.net;zuoxueliang0202@126.com
Share article
CLC Number:
CAI Juan, CHEN Zhiqiang, ZUO Xueliang. CircSMARCA5 inhibits glycolysis and suppresses proliferation and invasion of gastric cancer cells through miR-4295/PTEN axis[J]. China Oncology, 2022, 32(4): 298-308.
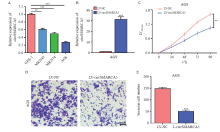
Fig. 1
CircSMARCA5 overexpression suppresses the proliferation and invasion of gastric cancer cells A: The expression levels of circSMARCA5 in three human gastric cancer cell lines and the normal human gastric mucosal epithelial cell line GES-1; B: RTFQ-PCR analysis results of circSMARCA5 in AGS cells after transfection of lentivirus overexpressing circSMARCA5; C: The proliferation of AGS cells after circSMARCA5 overexpression was detected by CCK-8 assays; D: The invasion of AGS cells after circSMARCA5 overexpression was examined by transwell assays; E: Invasion cell number of LV-NC and LV-circAMARCA5. **: P<0.01, compared with each other; ***: P<0.001, compared with each other."
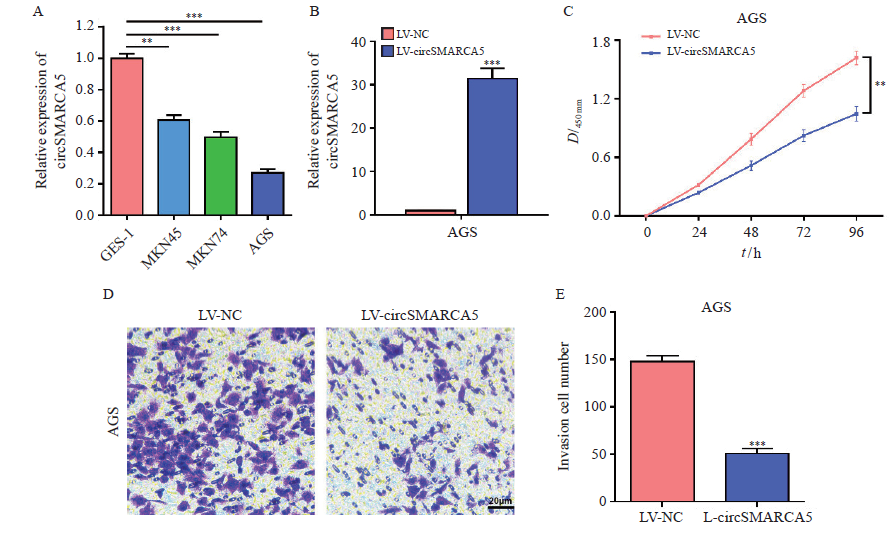
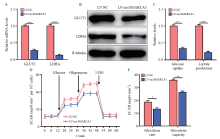
Fig. 2
CircSMARCA5 overexpression inhibits glycolysis of the gastric cancer cells A: The mRNA levels of GLUT1 and LDHA were examined by RTFQ-PCR in AGS cells with circSMARCA5 overexpression; B: The protein levels of GLUT1 and LDHA were assessed by Western blot in AGS cells with circSMARCA5 overexpression; C: The cellular glucose uptake and lactate production were detected in AGS cells with circSMARCA5 overexpression; D: Extracellular acidification rate (ECAR) was measured by Seahorse XF assays in AGS cells; E: ECAR data showed that upregulating circSMARCA5 significantly inhibited the glycolysis rate and glycolysis capacity in AGS cells. *: P<0.05, compared with each other; **: P<0.01, compared with each other; ***: P<0.001, compared with each other."
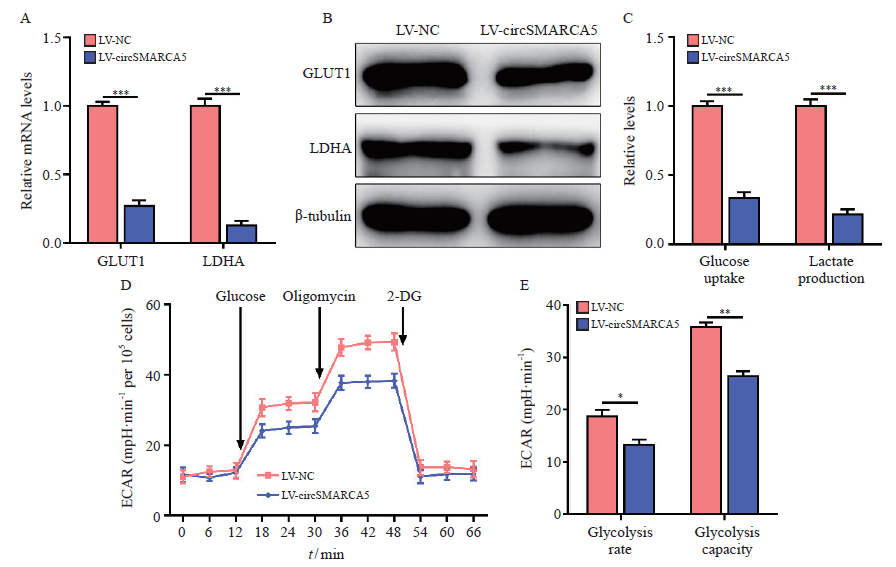
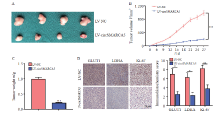
Fig. 3
CircSMARCA5 overexpression suppresses xenograft tumor growth in vivo A: Xenograft tumors were photographed; B: Growth curves of xenograft tumors were measured by tumor volume; C: Tumor weight was recorded; D: The expression levels of GLUT1, LDHA and Ki-67 proliferation index in xenograft tumors were analyzed by using immunohistochemistry. *: P<0.05, compared with each other; **: P<0.01, compared with each other; ***: P<0.001, compared with each other."
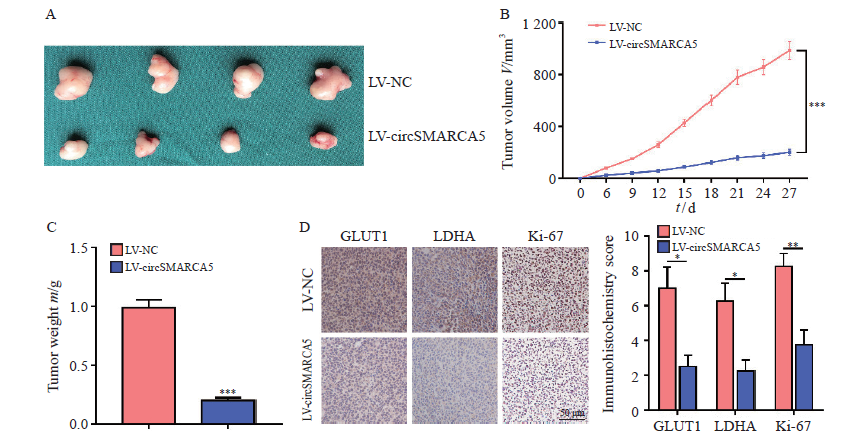
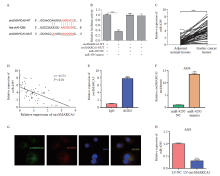
Fig. 4
CircSMARCA5 directly targets miR-4295 A: Putative binding sequence between circSMARCA5 and miR-4295; B: Luciferase activity was determined by dual luciferase reporter assays; C: The expression levels of miR-4295 in gastric cancer tissues and adjacent normal tissues were detected by RTFQ-PCR; D: Correlation analysis showed a negative correlation between circSMARCA5 and miR-4295 expression in gastric cancer tissues; E: RIP assays for AGO2 was conducted to detect the levels of endogenous circSMARCA5; F: Enrichment of circSMARCA5 in AGS cells transfected with miR-4295 NC or miR-4295 mimics; G: FISH results showed the colocalization of circSMARCA5 and miR-4295 in cytoplasm of gastric cancer cells; H: The expression of miR-4295 was detected by RTFQ-PCR in AGS cells with circSMARCA5 overexpression. ***: P<0.001, compared with each other."

Tab. 1
The clinicopathological features of 60 gastric cancer patients"
| Clinicopathological features | Case n (%) | Clinicopathological features | Case n (%) |
|---|---|---|---|
| Age/year | Lymph node metastasis | ||
| <60 | 21 (35.0) | Negative | 22 (36.7) |
| ≥60 | 39 (65.0) | Positive | 38 (63.3) |
| Gender | Vascular invasion | ||
| Female | 16 (26.7) | No | 29 (48.3) |
| Male | 44 (73.3) | Yes | 31 (51.7) |
| Tumor size D/cm | TNM staging | ||
| <5 | 41 (68.3) | Ⅰ/Ⅱ | 24 (40.0) |
| ≥5 | 19 (31.7) | Ⅲ | 36 (60.0) |
| Differentiation | |||
| Well/Moderate | 28 (46.7) | ||
| Poor | 32 (53.3) |
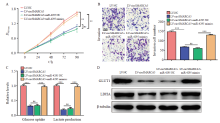
Fig. 5
CircSMARCA5 inhibits the proliferation and invasion of gastric cancer cells through targeting miR-4295 A: CCK-8 assay was performed to examine the cell proliferation of circSMARCA5-overexpressing cells with miR-4295 upregulation; B: Transwell assays were conducted to assess the invasive capability of circSMARCA5-overexpressing cells with miR-4295 upregulation; C: The cellular glucose uptake and lactate production were detected in circSMARCA5-overexpressing cells with miR-4295 upregulation; D: Western blot analysis showed the expression levels of GLUT1 and LDHA. **: P<0.01, compared with each other; ***: P<0.001, compared with each other; ns: No significance."
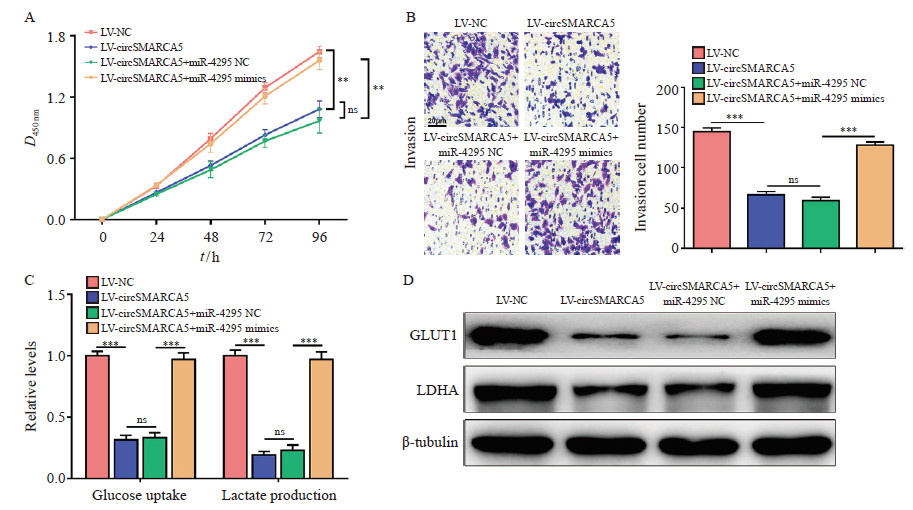
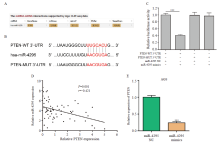
Fig. 6
PTEN was a direct target gene of miR-4295 A: ENCORI database prediction indicated that PTEN was a potential direct target gene of miR-4295; B: Putative binding sequence between miR-4295 and PTEN; C: Luciferase activity was determined using dual luciferase reporter assays; D: A negative correlation between miR-4295 and PTEN expression level was demonstrated in gastric cancer tissues; E: The expression level of PTEN was detected by RTFQ-PCR in AGS cells with miR-4295 overexpression. ***P<0.001."
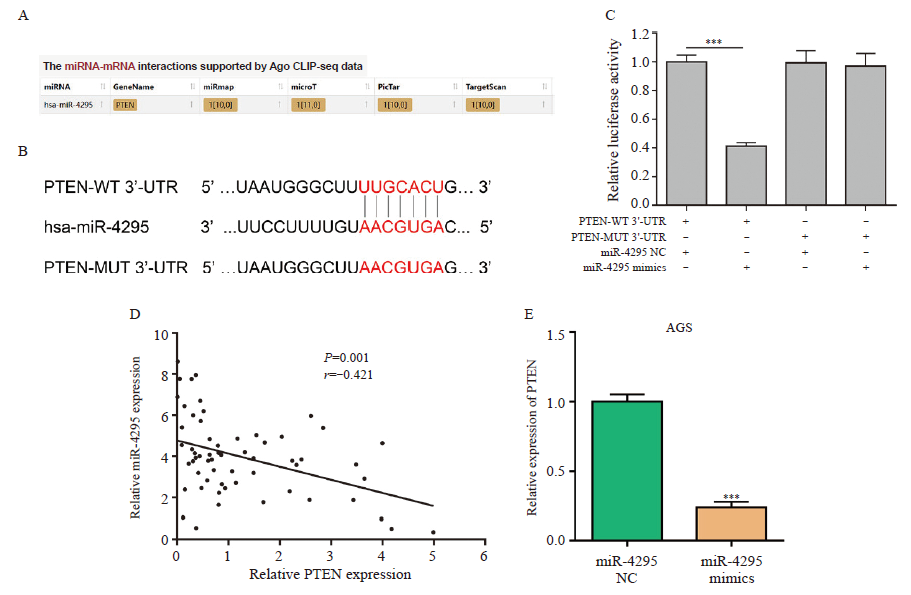
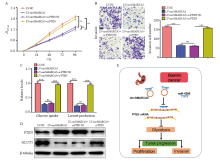
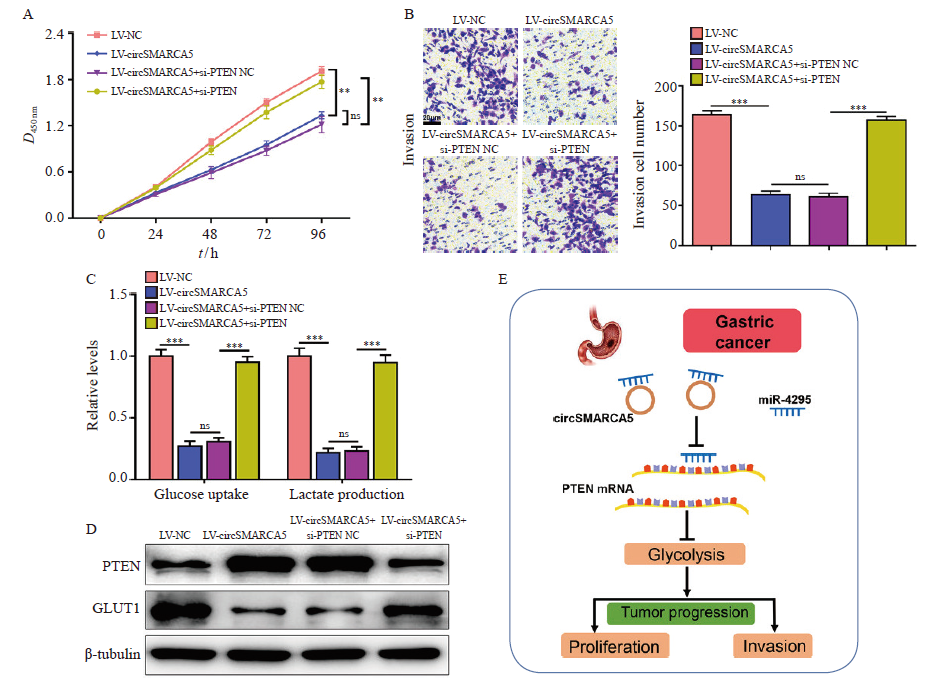
Fig. 7
CircSMARCA5 inhibits the proliferation and invasion of gastric cancer cells through miR-4295/PTEN axis A: CCK-8 assays were performed to examine the cell proliferation of circSMARCA5-overexpressing cells with PTEN knockdown; B: Transwell assays were conducted to assess the invasive capability of circSMARCA5-overexpressing cells with PTEN knockdown; C: The cellular glucose uptake and lactate production were detected in circSMARCA5-overexpressing cells with PTEN knockdown; D: Western blot analysis showing the expression levels of PTEN and GLUT1; E: Schematic diagram demonstrating the molecular mechanisms underlying circSMARCA5 in gastric cancer. **: P<0.01, compared with each other; ***: P<0.001, compared with each other; ns: No significance."
| [1] |
SUNG H,, FERLAY J,, SIEGEL R L, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71(3): 209-249.
doi: 10.3322/caac.21660 |
| [2] |
KIM E,, KIM Y K,, LEE S J V. Emerging functions of circular RNA in aging[J]. Trends Genet, 2021, 37(9): 819-829.
doi: 10.1016/j.tig.2021.04.014 |
| [3] |
LU J,, WANG Y H,, YOON C, et al. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis[J]. Cancer Lett, 2020, 471: 38-48.
doi: 10.1016/j.canlet.2019.11.038 |
| [4] |
LI J F,, HUANG C C,, ZOU Y F, et al. CircTLK1 promotes the proliferation and metastasis of renal cell carcinoma by sponging miR-136-5p[J]. Mol Cancer, 2020, 19(1): 103.
doi: 10.1186/s12943-020-01225-2 |
| [5] | CAI J,, CHEN Z Q,, ZUO X L. circSMARCA 5 functions as a diagnostic and prognostic biomarker for gastric cancer[J]. Dis Markers, 2019, 2019: 2473652. |
| [6] |
YU T,, WANG Y F,, FAN Y, et al. CircRNAs in cancer metabolism: a review[J]. J Hematol Oncol, 2019, 12(1): 90.
doi: 10.1186/s13045-019-0776-8 |
| [7] |
LI J,, SUN D,, PU W C, et al. Circular RNAs in cancer: biogenesis, function, and clinical significance[J]. Trends Cancer, 2020, 6(4): 319-336.
doi: 10.1016/j.trecan.2020.01.012 |
| [8] |
HE A T,, LIU J L,, LI F Y, et al. Targeting circular RNAs as a therapeutic approach: current strategies and challenges[J]. Signal Transduct Target Ther, 2021, 6(1): 185.
doi: 10.1038/s41392-021-00569-5 |
| [9] |
WANG S M,, ZHANG K,, TAN S Y, et al. Circular RNAs in body fluids as cancer biomarkers: the new frontier of liquid biopsies[J]. Mol Cancer, 2021, 20(1): 13.
doi: 10.1186/s12943-020-01298-z |
| [10] | BEILERLI A,, GAREEV I,, BEYLERLI O, et al. Circular RNAs as biomarkers and therapeutic targets in cancer[J]. Semin Cancer Biol, 2021: S1044-S579X(21)00004-3. |
| [11] |
YU J,, XU Q G,, WANG Z G, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma[J]. J Hepatol, 2018, 68(6): 1214-1227.
doi: 10.1016/j.jhep.2018.01.012 |
| [12] | BARBAGALLO D,, CAPONNETTO A,, CIRNIGLIARO M, et al. CircSMARCA 5 inhibits migration of glioblastoma multiforme cells by regulating a molecular axis involving splicing factors SRSF1/SRSF3/PTB[J]. Int J Mol Sci, 2018, 19(2): E480. |
| [13] |
VANDER HEIDEN M G,, CANTLEY L C,, THOMPSON C B. Understanding the Warburg effect: the metabolic requirements of cell proliferation[J]. Science, 2009, 324(5930): 1029-1033.
doi: 10.1126/science.1160809 |
| [14] |
EL HASSOUNI B,, GRANCHI C,, VALLÉS-MARTÍ A, et al. The dichotomous role of the glycolytic metabolism pathway in cancer metastasis: interplay with the complex tumor microenvironment and novel therapeutic strategies[J]. Semin Cancer Biol, 2020, 60: 238-248.
doi: 10.1016/j.semcancer.2019.08.025 |
| [15] |
MENG F Y,, WU L,, DONG L, et al. EGFL9 promotes breast cancer metastasis by inducing cMET activation and metabolic reprogramming[J]. Nat Commun, 2019, 10(1): 5033.
doi: 10.1038/s41467-019-13034-3 |
| [16] |
PU Z C,, XU M D,, YUAN X L, et al. Circular RNA circCUL3 accelerates the Warburg effect progression of gastric cancer through regulating the STAT3/HK2 axis[J]. Mol Ther Nucleic Acids, 2020, 22: 310-318.
doi: 10.1016/j.omtn.2020.08.023 |
| [17] |
YUAN Q G,, ZHANG Y,, LI J H, et al. High expression of microRNA-4295 contributes to cell proliferation and invasion of pancreatic ductal adenocarcinoma by the down-regulation of glypican-5[J]. Biochem Biophys Res Commun, 2018, 497(1): 73-79.
doi: 10.1016/j.bbrc.2018.02.023 |
| [18] | NAN Y H,, WANG J,, WANG Y, et al. Erratum: miR-4295 promotes cell growth in bladder cancer by targeting BTG1[J]. Am J Transl Res, 2017, 9(4): 1960. |
| [19] | YAN R,, LI K,, YUAN D W, et al. Downregulation of microRNA-4295 enhances cisplatin-induced gastric cancer cell apoptosis through the EGFR/PI3K/Akt signaling pathway by targeting LRIG1[J]. Int J Oncol, 2018, 53(6): 2566-2578. |
| [20] |
YAN X B,, WANG T,, WANG J. Circ_0016760 acts as a sponge of microRNA-4295 to enhance E2F transcription factor 3 expression and facilitates cell proliferation and glycolysis in non-small cell lung cancer[J]. Cancer Biother Radiopharm, 2022, 37(2): 147-158.
doi: 10.1089/cbr.2020.3621 |
| [21] |
CHEN C Y,, CHEN J Y,, HE L N, et al. PTEN: tumor suppressor and metabolic regulator[J]. Front Endocrinol (Lausanne), 2018, 9: 338.
doi: 10.3389/fendo.2018.00338 |
| [22] |
ORTEGA-MOLINA A,, SERRANO M. PTEN in cancer, metabolism, and aging[J]. Trends Endocrinol Metab, 2013, 24(4): 184-189.
doi: 10.1016/j.tem.2012.11.002 |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
沪ICP备12009617
Powered by Beijing Magtech Co. Ltd
