Welcome to China Oncology,
China Oncology ›› 2024, Vol. 34 ›› Issue (5): 460-472.doi: 10.19401/j.cnki.1007-3639.2024.05.003
• Article • Previous Articles Next Articles
ZHOU Xueqin( ), LUAN Yanchao(
), LUAN Yanchao( ), ZHAO Li, RONG Chaochao, YANG Na
), ZHAO Li, RONG Chaochao, YANG Na
Received:2023-11-17
Revised:2024-04-22
Online:2024-05-30
Published:2024-06-07
Contact:
LUAN Yanchao
Share article
CLC Number:
ZHOU Xueqin, LUAN Yanchao, ZHAO Li, RONG Chaochao, YANG Na. Expression of CDC20 in lung adenocarcinoma tissues and its effect on the proliferation and invasion of lung adenocarcinoma cells[J]. China Oncology, 2024, 34(5): 460-472.
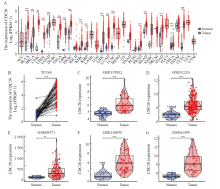
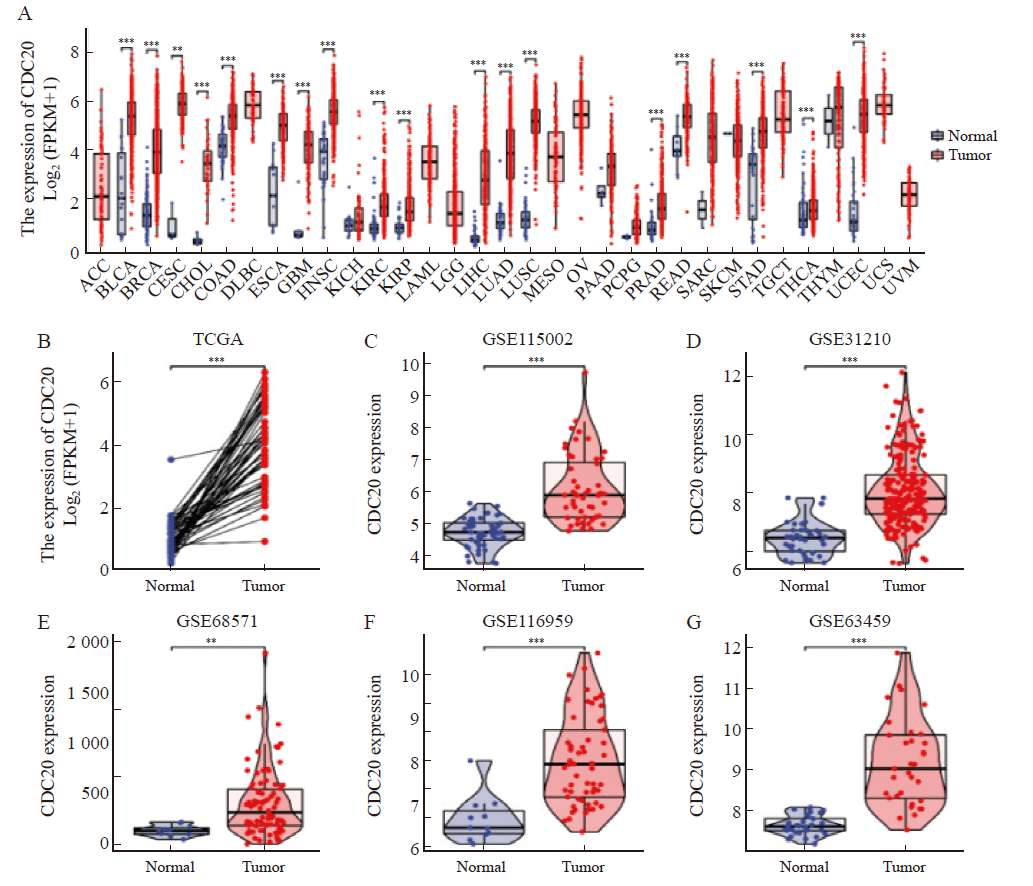
Fig. 1
CDC20 is highly expressed in multiple tumor types A: Differential expression of CDC20 in pan-cancer samples from the TCGA database; B: CDC20 expression in lung adenocarcinoma and matched normal tissues in the TCGA databases; C-G: The CDC20 mRNA expression between lung adenocarcinoma and normal tissues based on data from GSE115002 (C), GSE31210 (D), GSE68571 (E),GSE116959(F) and GSE116959(G) dataset. **: P<0.01, compared with normal. ***: P<0.001, compared with normal."
Tab. 1
Relationship between expression of CDC20 and clinicopathological features in lung adenocarcinoma [n (%)]"
| Categories | CDC20 expression | χ2 value | P value | |
|---|---|---|---|---|
| High expression (n=63) | Low expression (n=55) | |||
| Age/year | 0.034 | 0.854 | ||
| <60 | 32 (27.1) | 27 (22.9) | ||
| ≥60 | 31 (26.3) | 28 (23.7) | ||
| Gender | 0.025 | 0.873 | ||
| Male | 33 (28) | 28 (23.7) | ||
| Female | 30 (25.4) | 27 (22.9) | ||
| Clinical stage | 10.926 | < 0.001 | ||
| Ⅰ-Ⅱ | 32 (27.1) | 44 (37.3) | ||
| Ⅲ-Ⅳ | 31 (26.3) | 11 (9.3) | ||
| T classification | 9.721 | 0.002 | ||
| T1-T2 | 36 (30.5) | 46 (39) | ||
| T3-T4 | 27 (22.9) | 9 (7.6) | ||
| N classification | 8.224 | 0.004 | ||
| No | 14 (11.9) | 26 (22) | ||
| Yes | 49 (41.5) | 29 (24.6) | ||
| M classification | 2.329 | 0.127 | ||
| No | 56 (47.5) | 53 (44.9) | ||
| Yes | 7 (5.9) | 2 (1.7) | ||
| Degree of differentiation | 21.32 | < 0.001 | ||
| Medium/low | 35 (29.7) | 8 (6.8) | ||
| High | 28 (23.7) | 47 (39.8) | ||
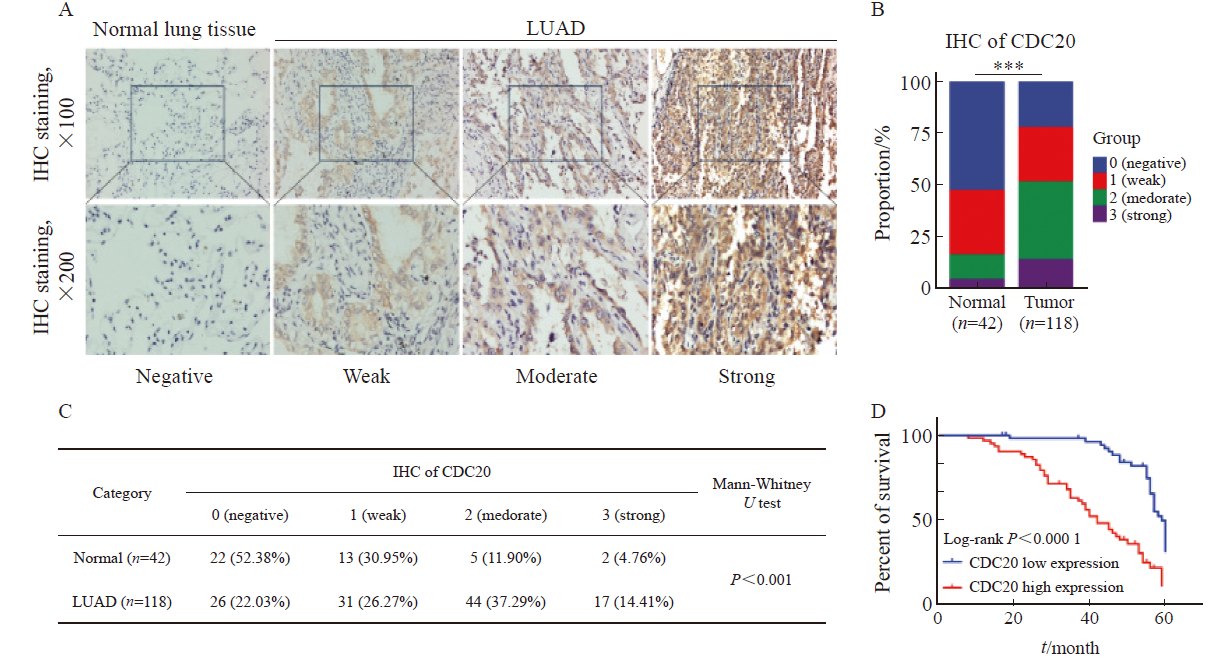
Fig. 4
Correlation between CDC20 expression and prognosis in lung adenocarcinoma A: IHC for detecting CDC20 expression in lung adenocarcinoma tissue and Normal lung tissue; B, C: Statistical analysis of tissue IHC scores; D: Kaplan-Meier analysis of CDC20 expression and OS based on lung adenocarcinoma tissue. ***: P<0.001, compared with normal."
Tab. 2
COX regression analysis of clinicopathological features and OS rate in lung adenocarcinoma"
| Characteristic | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | HR (95% CI) | P value | ||
| Age/year | 1.148 (0.723-1.824) | 0.558 | |||
| Gender | 0.853 (0.535-1.362) | 0.506 | |||
| Clinical stage | 0.167 (0.102-0.275) | < 0.001 | 0.383 (0.226-0.651) | <0.001 | |
| T classification | 4.217 (2.567-6.928) | < 0.001 | 1.428 (0.860-2.373) | 0.169 | |
| N classification | 0.198 (0.112-0.351) | < 0.001 | 0.347 (0.191-0.627) | <0.001 | |
| M classification | 80.72 (19.01-342.72) | < 0.001 | 32.50 (7.70-137.23) | <0.001 | |
| Degree of differentiation | 0.491 (0.305-0.792) | 0.004 | 0.801 (0.483-1.328) | 0.389 | |
| CDC20 expression | 0.246 (0.144-0.419) | < 0.001 | 0.249 (0.141-0.440) | <0.001 | |

Fig. 5
Expression levels of CDC20 mRNA and protein in lung adenocarcinoma cells in different groups A: Expression levels of CDC20 mRNA and protein in BEAS-2B, A549 and H1299 cells; B: Relative expression levels of CDC20 mRNA and protein in A549 cells with CDC20 knockdown; C: Relative expression levels of CDC20 mRNA and protein in H1299 cells with CDC20 knockdown. ***: P <0.001, compared with BEAS-2B or siNC."
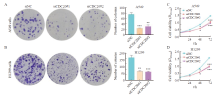
Fig. 6
Effect of CDC20 silencing on the proliferation ability of A549 and H1299 cells A, B: Colony formation assay showed that knockdown of CDC20 inhibited the proliferation of A549 and H1299 cells; C, D: CCK-8 assay showed that knockdown of CDC20 inhibited the proliferation of A549 and H1299 cells. *: P<0.05, compared with siNC; **: P<0.01, compared with siNC; ***: P<0.001, compared with siNC."
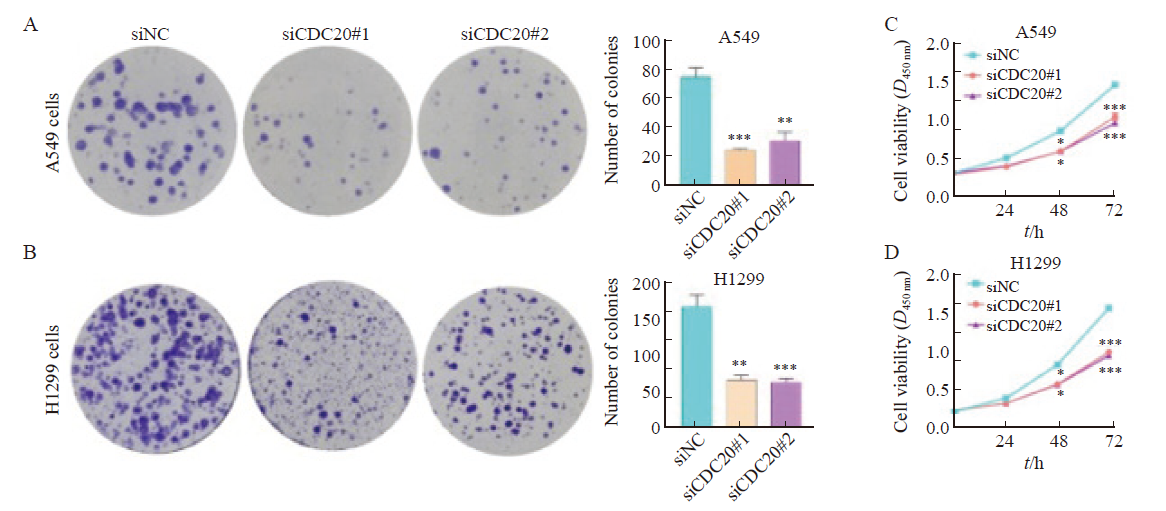
Fig. 7
Effect of CDC20 silencing on the migration and invasion of A549 and H1299 cells A, B: Effect of CDC20 silencing on the migration ability of A549 and H1299 cells; C, D: Effect of CDC20 silencing on the invasion of A549 and H1299 cells. **: P<0.01, compared with siNC; ***: P<0.001, compared with siNC."
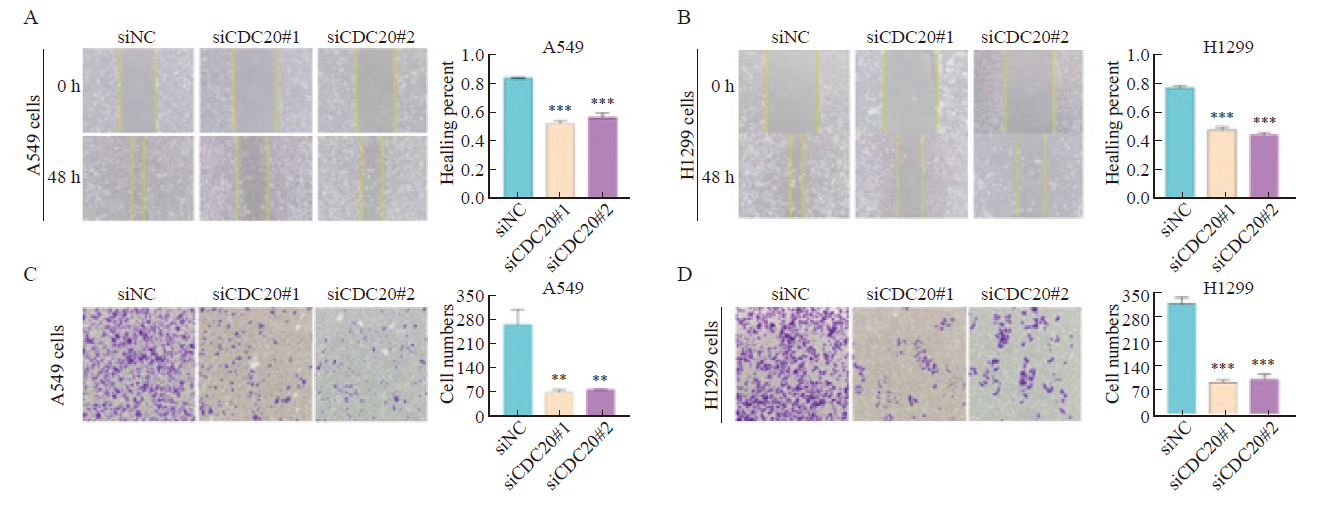
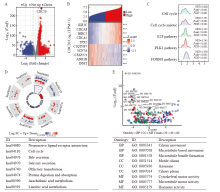
Fig. 8
Results of DEG and functional enrichment analysis A: Volcano plot of DEG (red: upregulation; blue: downregulation); B: Heatmap of the correlation between CDC20 expression and the top 5 DEG;C: Results of GSEA enrichment analysis of DEG; D: KEGG pathway analysis; E: GO functional annotation analysis of biological process (BP), cellular component (CC) and molecular function (MF)."

| [1] | SIEGEL R L, MILLER K D, JEMAL A. Cancer statistics, 2016[J]. CA Cancer J Clin, 2016, 66(1): 7-30. |
| [2] | CASCONE T, FRADETTE J, PRADHAN M, et al. Tumor immunology and immunotherapy of non-small cell lung cancer[J]. Cold Spring Harb Perspect Med, 2022, 12(5): a037895. |
| [3] | CHEN P X, LIU Y H, WEN Y K, et al. Non-small cell lung cancer in China[J]. Cancer Commun, 2022, 42(10): 937-970. |
| [4] |
SINGH S S, DAHAL A, SHRESTHA L, et al. Genotype driven therapy for non-small cell lung cancer: resistance, pan inhibitors and immunotherapy[J]. Curr Med Chem, 2020, 27(32): 5274-5316.
doi: 10.2174/0929867326666190222183219 pmid: 30854949 |
| [5] | CHENG S J, CASTILLO V, SLIVA D. CDC20 associated with cancer metastasis and novel mushroom-derived CDC20 inhibitors with antimetastatic activity[J]. Int J Oncol, 2019, 54(6): 2250-2256. |
| [6] |
CHI J J, LI H C, ZHOU Z, et al. A novel strategy to block mitotic progression for targeted therapy[J]. EBioMedicine, 2019, 49: 40-54.
doi: S2352-3964(19)30677-2 pmid: 31669221 |
| [7] |
WANG J J, ZHOU F G, LI Y, et al. Cdc20 overexpression is involved in temozolomide-resistant glioma cells with epithelial-mesenchymal transition[J]. Cell Cycle, 2017, 16(24): 2355-2365.
doi: 10.1080/15384101.2017.1388972 pmid: 29108461 |
| [8] | MA X, ZHOU L, ZHENG S S. Transcriptome analysis revealed key prognostic genes and microRNAs in hepatocellular carcinoma[J]. PeerJ, 2020, 8: e8930. |
| [9] | LOVE M I, HUBER W, ANDERS S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biol, 2014, 15(12): 550. |
| [10] |
LIU J F, LICHTENBERG T, HOADLEY K A, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics[J]. Cell, 2018, 173(2): 400-416.e11.
doi: S0092-8674(18)30229-0 pmid: 29625055 |
| [11] |
YU G C, WANG L G, HAN Y Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters[J]. OMICS, 2012, 16(5): 284-287.
doi: 10.1089/omi.2011.0118 pmid: 22455463 |
| [12] | GIOPANOU I, KANELLAKIS N I, GIANNOU A D, et al. Osteopontin drives KRAS-mutant lung adenocarcinoma[J]. Carcinogenesis, 2020, 41(8): 1134-1144. |
| [13] |
贾利晴, 葛小路, 姜琳, 等. LncRNA PKD2-2-3对肺腺癌细胞增殖、克隆形成、迁移及侵袭能力的影响[J]. 中国癌症杂志, 2023, 33(8): 717-725.
doi: 10.19401/j.cnki.1007-3639.2023.08.001 |
| [14] | JIA L Q, GE X L, JIANG L, et al. The impact of lncRNA PKD2-2-3 on the proliferation, colony formation, migration, and invasion abilities of lung adenocarcinoma cells[J]. China Oncol, 2023, 33(8): 717-725. |
| [15] |
WU Q W, YU L L, LIN X Q, et al. Combination of serum miRNAs with serum exosomal miRNAs in early diagnosis for non-small-cell lung cancer[J]. Cancer Manag Res, 2020, 12: 485-495.
doi: 10.2147/CMAR.S232383 pmid: 32021461 |
| [16] | 刘晓丽, 柴文君, 孙磊, 等. 肺腺癌中可变剪接调控因子KHSRP调控的差异剪接基因分析[J]. 中国癌症杂志, 2023, 33(07): 637-645. |
| [17] | LIU X L, CHAI W J, SUN L, et al. Analysis of differentially spliced genes regulated by the alternative splicing factor KHSRP in lung adenocarcinoma[J]. China Oncol, 2023, 33(7): 637-645. |
| [18] |
CUADRADO M, GARZÓN J, MORENO S, et al. Efficient terminal erythroid differentiation requires the APC/C cofactor Cdh1 to limit replicative stress in erythroblasts[J]. Sci Rep, 2022, 12(1):10489.
doi: 10.1038/s41598-022-14331-6 pmid: 35729193 |
| [19] | KIM J H, PATEL R. Mad2B forms a complex with Cdc20, Cdc27, Rev3 and Rev1 in response to cisplatin-induced DNA damage[J]. Korean J Physiol Pharmacol, 2023, 27(5): 427-436. |
| [20] | ZHANG Y, XUE Y B, LI H, et al. Inhibition of cell survival by curcumin is associated with downregulation of cell division cycle 20 (Cdc20) in pancreatic cancer cells[J]. Nutrients, 2017, 9(2): 109. |
| [21] |
PARMAR M B, K C R B, LÖBENBERG R, et al. Additive polyplexes to undertake siRNA therapy against CDC20 and survivin in breast cancer cells[J]. Biomacromolecules, 2018, 19(11): 4193-4206.
doi: 10.1021/acs.biomac.8b00918 pmid: 30222931 |
| [22] |
LI K, MAO Y H, LU L, et al. Silencing of CDC20 suppresses metastatic castration-resistant prostate cancer growth and enhances chemosensitivity to docetaxel[J]. Int J Oncol, 2016, 49(4): 1679-1685.
doi: 10.3892/ijo.2016.3671 pmid: 27633058 |
| [23] | LI J X, WANG Y C, WANG X, et al. CDK1 and CDC20 overexpression in patients with colorectal cancer are associated with poor prognosis: evidence from integrated bioinformatics analysis[J]. World J Surg Oncol, 2020, 18(1): 50. |
| [24] | YANG G, WANG G, XIONG Y F, et al. CDC20 promotes the progression of hepatocellular carcinoma by regulating epithelial-mesenchymal transition[J]. Mol Med Rep, 2021, 24(1): 483. |
| [25] | ZHANG Y Q, LI J Y, YI K K, et al. Elevated signature of a gene module coexpressed with CDC20 marks genomic instability in glioma[J]. Proc Natl Acad Sci U S A, 2019, 116(14): 6975-6984. |
| [26] | WANG L X, ZHANG J F, WAN L X, et al. Targeting CDC20 as a novel cancer therapeutic strategy[J]. Pharmacol Ther, 2015, 151: 141-151. |
| [27] |
WANG L X, YANG C L, CHU M, et al. Cdc20 induces the radioresistance of bladder cancer cells by targeting FoxO1 degradation[J]. Cancer Lett, 2021, 500: 172-181.
doi: 10.1016/j.canlet.2020.11.052 pmid: 33290869 |
| [28] | CHEN Z H, JING Y J, YU J B, et al. ESRP1 induces cervical cancer cell G1-phase arrest via regulating cyclin A2 mRNA stability[J]. Int J Mol Sci, 2019, 20(15): 3705. |
| [29] | AMADOR V, GE S, SANTAMARÍA P G, et al. APC/C (CDC20) controls the ubiquitin-mediated degradation of p21 in prometaphase[J]. Mol Cell, 2007, 27(3): 462-473. |
| [30] | ZHENG Y C, SHI Y, YU S, et al. GTSE1, CDC20, PCNA, and MCM6 synergistically affect regulations in cell cycle and indicate poor prognosis in liver cancer[J]. Anal Cell Pathol, 2019, 2019: 1038069. |
| [31] |
KAPANIDOU M, CURTIS N L, BOLANOS-GARCIA V M. CDC20: at the crossroads between chromosome segregation and mitotic exit[J]. Trends Biochem Sci, 2017, 42(3): 193-205.
doi: S0968-0004(16)30209-2 pmid: 28202332 |
| [32] | CHU Z W, ZHANG X Y, LI Q Y, et al. CDC20 contributes to the development of human cutaneous squamous cell carcinoma through the Wnt/β-catenin signaling pathway[J]. Int J Oncol, 2019, 54(5): 1534-1544. |
| [33] |
LI J, KARKI A, HODGES K B, et al. Cotargeting polo-like kinase 1 and the Wnt/β-catenin signaling pathway in castration-resistant prostate cancer[J]. Mol Cell Biol, 2015, 35(24): 4185-4198.
doi: 10.1128/MCB.00825-15 pmid: 26438599 |
| [34] | KONG J J, WANG X T, WANG J L, et al. Silencing of RAB42 down-regulated PD-L1 expression to inhibit the immune escape of hepatocellular carcinoma cells through inhibiting the E2F signaling pathway[J]. Cell Signal, 2023, 108: 110692. |
| [35] | ZHANG Y, LYU L, WANG W, et al. High expression of E2F transcription factors 7: an independent predictor of poor prognosis in patients with lung adenocarcinoma[J]. Medicine, 2022, 101(33): e29253. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
沪ICP备12009617
Powered by Beijing Magtech Co. Ltd
