Welcome to China Oncology,
China Oncology ›› 2024, Vol. 34 ›› Issue (11): 987-997.doi: 10.19401/j.cnki.1007-3639.2024.11.001
• Article • Previous Articles Next Articles
WU Zhibai1( ), XU Guiqin1, ZHANG Li1, YANG Zhaojuan1, LIU Yun1, JIAO Kun1,2, CHEN Zehong1, XU Chen1, ZUO You1, ZHENG Ningqian1, YE Zhiqian1, LIU Yongzhong1(
), XU Guiqin1, ZHANG Li1, YANG Zhaojuan1, LIU Yun1, JIAO Kun1,2, CHEN Zehong1, XU Chen1, ZUO You1, ZHENG Ningqian1, YE Zhiqian1, LIU Yongzhong1( )
)
Received:2024-06-26
Revised:2024-09-12
Online:2024-11-30
Published:2024-12-11
Contact:
LIU Yongzhong
Share article
CLC Number:
WU Zhibai, XU Guiqin, ZHANG Li, YANG Zhaojuan, LIU Yun, JIAO Kun, CHEN Zehong, XU Chen, ZUO You, ZHENG Ningqian, YE Zhiqian, LIU Yongzhong. Mechanism study of KCMF1 promoting proliferation and NF-κB signaling transduction in colorectal cancer cells[J]. China Oncology, 2024, 34(11): 987-997.
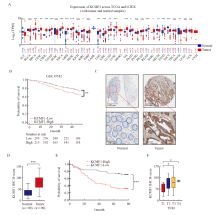
Fig. 1
High expression of KCMF1 is associated with poor prognosis of patients with CRC A: mRNA levels of KCMF1 in different types of tumor and normal tissues from the TCGA and GTEx databases. TPM: Transcripts per million; NS: No significance; *: P<0.05, assessed by Student’s t-tests; **: P<0.01, assessed by Student’s t-tests; ***: P<0.001, assessed by Student’s t-tests. B: The correlation between KCMF1 expression and the prognosis of CRC patients in GSE39582 dataset from the GEO database. **: P<0.01, assessed by log-rank tests. C: Representative images of immunohistochemical staining for KCMF1 in CRC tissues and their paired adjacent tissues. D: Immunohistochemical staining scores in the 90 colorectal tumor tissues and their paired adjacent tissues. ***: P<0.001, assessed by Student’s t-tests. E: The survival probability of 90 patients with high or low expression of KCMF1 in colorectal tumors; The KCMF1 scores are ranked by High and Low, with KCMF1-High group (n=45) and KCMF1-Low group (n=45). ***: P<0.001, assessed by log-rank tests. F: The correlation between KCMF1 expression and TNM stage in patients with CRC. *: P<0.05, assessed by Student-Newman-Keuls."
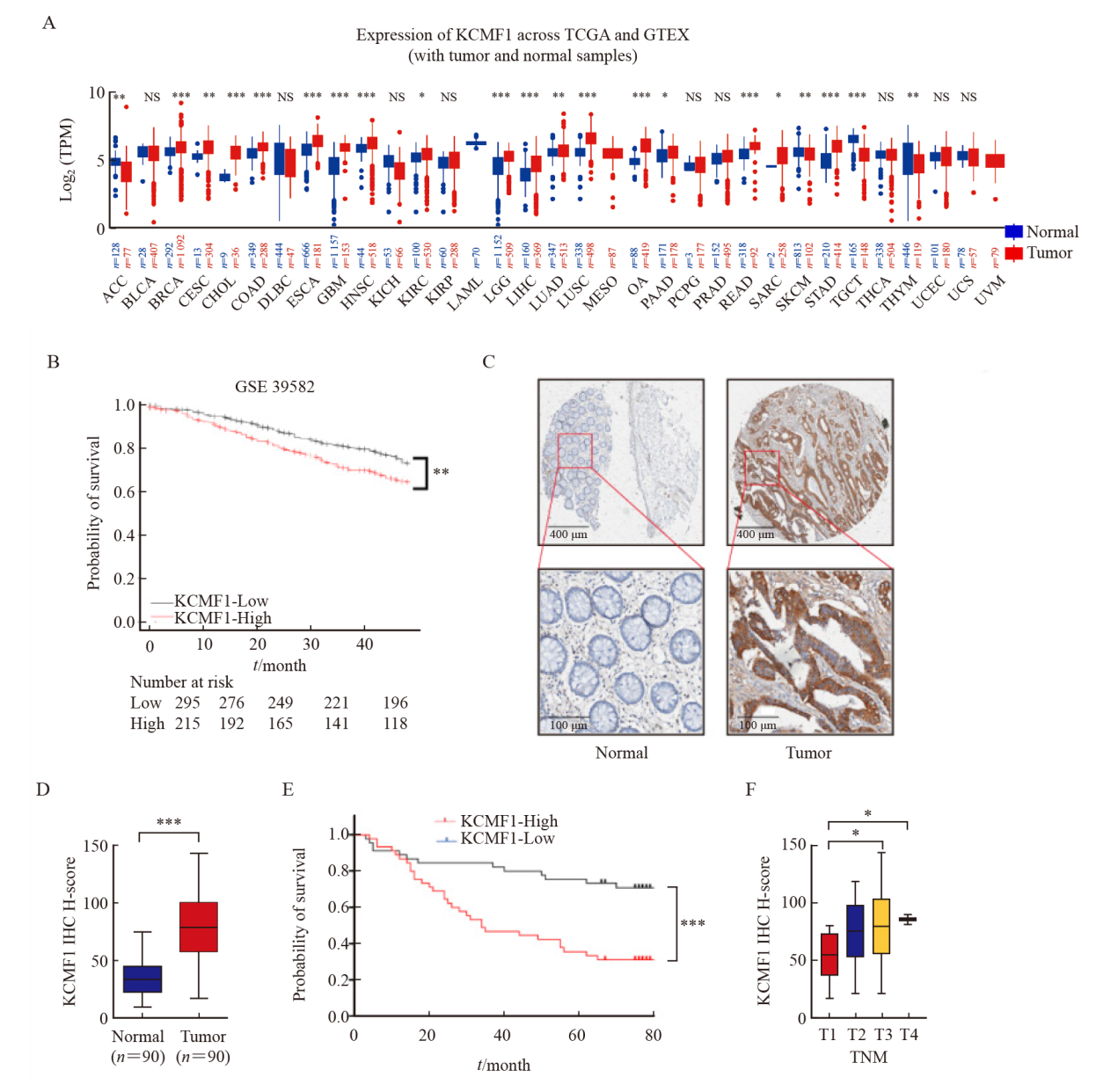

Fig. 2
Effects of KCMF1 knockdown on the proliferation of CRC cells in vitro A: Expression level of KCMF1 protein in HCT116 and HCT15 cells with or without shKCMF1 expression. B, C: The proliferation of HCT116 and HCT15 cells with or without shKCMF1 expression was assessed by MTT assay (B) and colony formation assay (C). ***: P<0.001, assessed by Student’s t-tests. D: Expression levels of the apoptosis-related proteins in HCT116 and HCT15 cells with or without shKCMF1. E: Flow cytometry analysis of the percentage of apoptotic cells in the indicated cells treated with or without Z-VAD-FMK (20 µm) for 48 h. NS: No significance; ***; P<0.001, assessed by Student-Newman-Keuls. F: Expression levels of the cell cycle-related proteins in HCT116 and HCT15 cells with or without shKCMF1. G: Flow cytometry analysis of the cell cycle distribution of HCT116 and HCT15 cells with or without shKCMF1. **: P<0.01, assessed by Student-Newman-Keuls; ***: P<0.001, assessed by Student-Newman-Keuls."
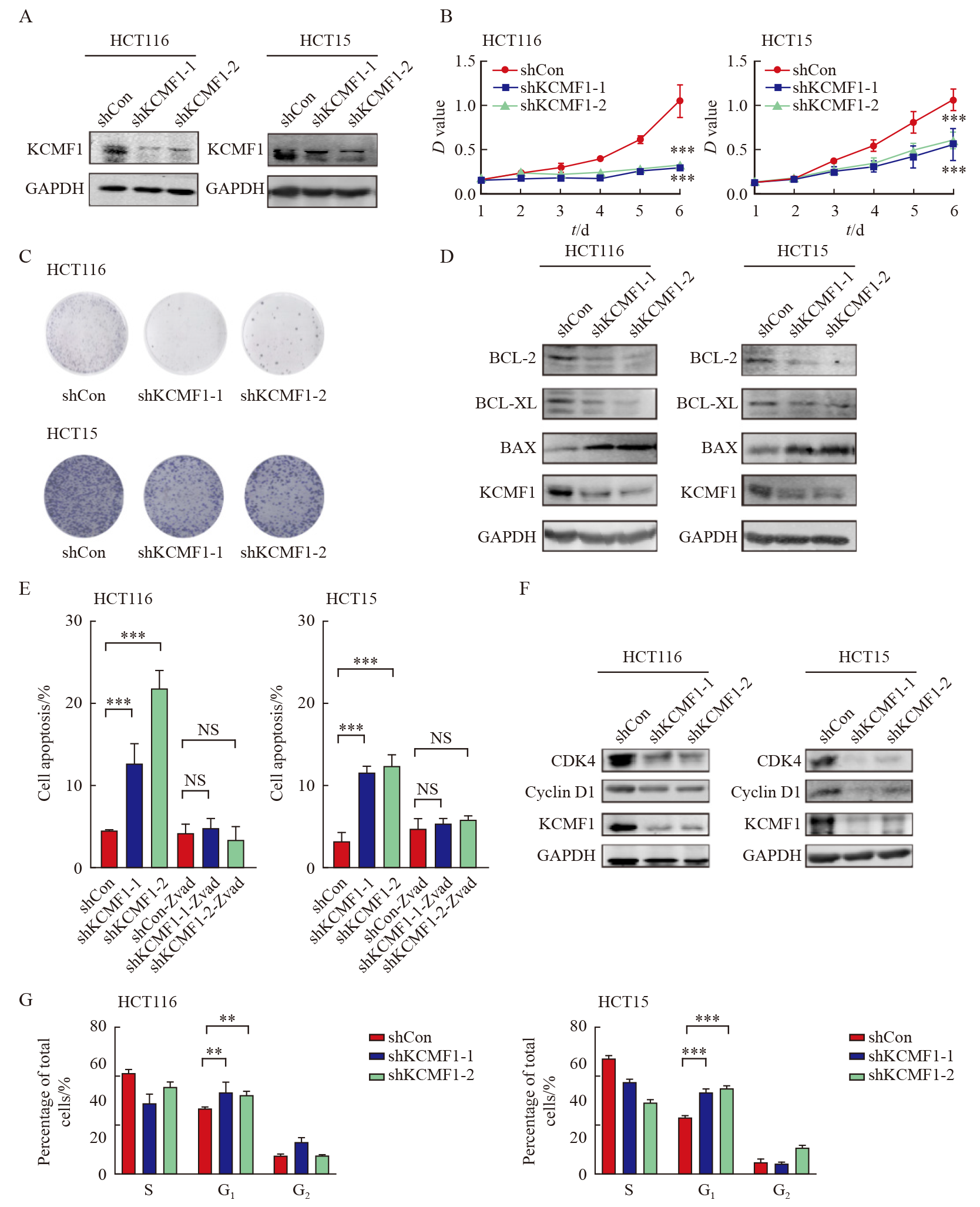

Fig. 3
Effects of KCMF1 knockdown on the signaling pathways in human CRC cells A: Volcano plots show differentially expressed genes affected by KCMF1 knockdown in HCT116 cells. The significant different genes are determined with the following criteria: |Log2 FC|>1 and P<0.05. FC: Fold change. B: GO analysis of the down-regulated genes in HCT116 cells with KCMF1 knockdown. P<0.05 and Log2 FC≤-1 were used as the criteria to define down-regulated genes. C: Enrichment analysis of the KCMF1-regulated genes with HALLMARK gene signatures, the significantly enriched signatures were ranked by P value, the bar plot shows the top 10 signatures. D: GSEA of the transcriptional profiles of KCMF1-koncokdown HCT116 cells and control cells with the TNFα-NF-κB signature. E: Spearman correlation analysis was performed between BIOCARTA_NFKB_PATHWAY and KCMF1 expression based on TCGA database cohort. F: The TCGA cohort were classified into four groups by both KCMF1 expression and NF-κB activity and subjected to Kaplan-Meier overall survival analysis. The activity of NF-κB signaling was determined by the GSVA score of the gene set BIOCARTA_NFKB_SIGNALING. **: P<0.01, assessed by log-rank tests."
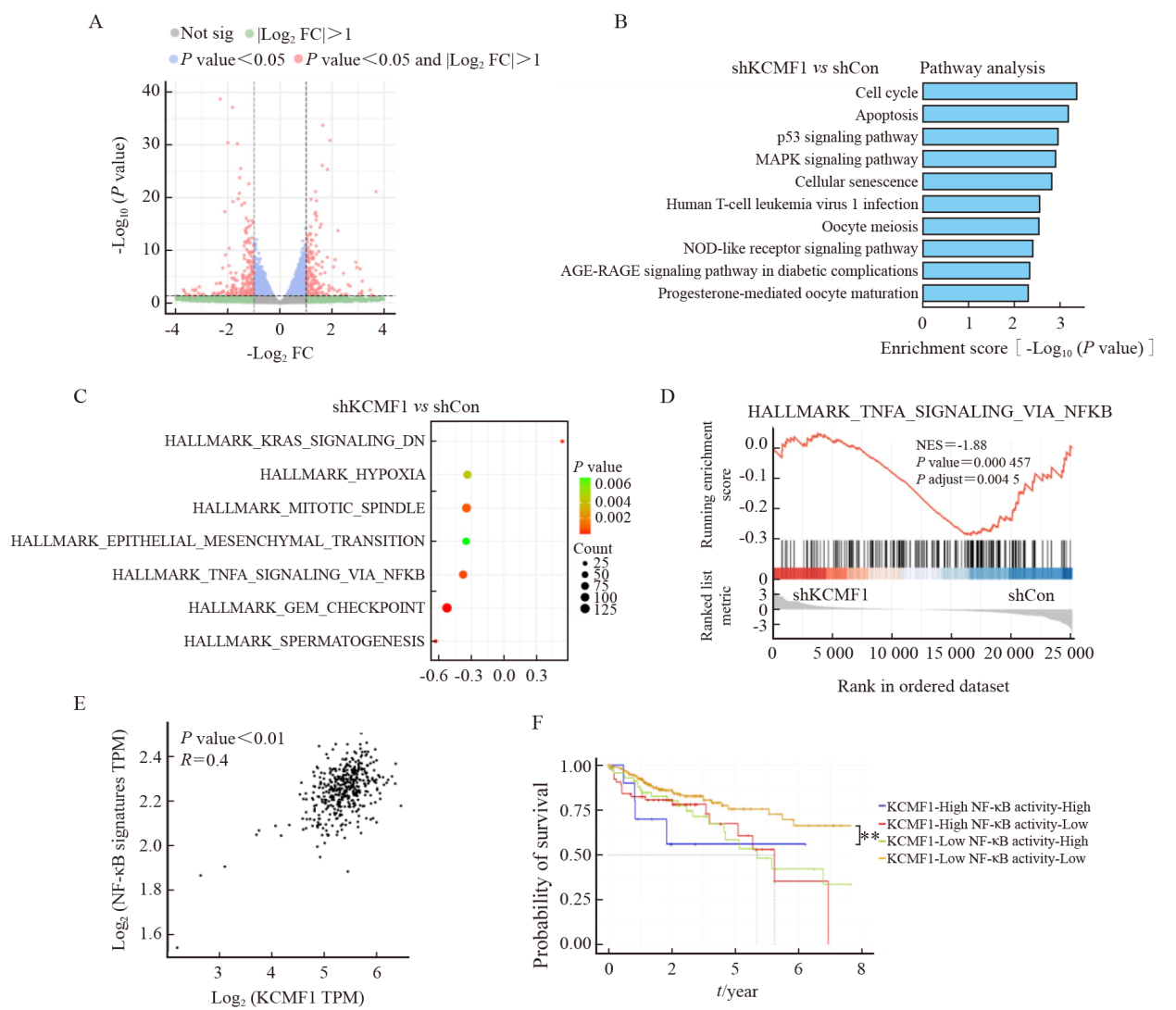

Fig. 4
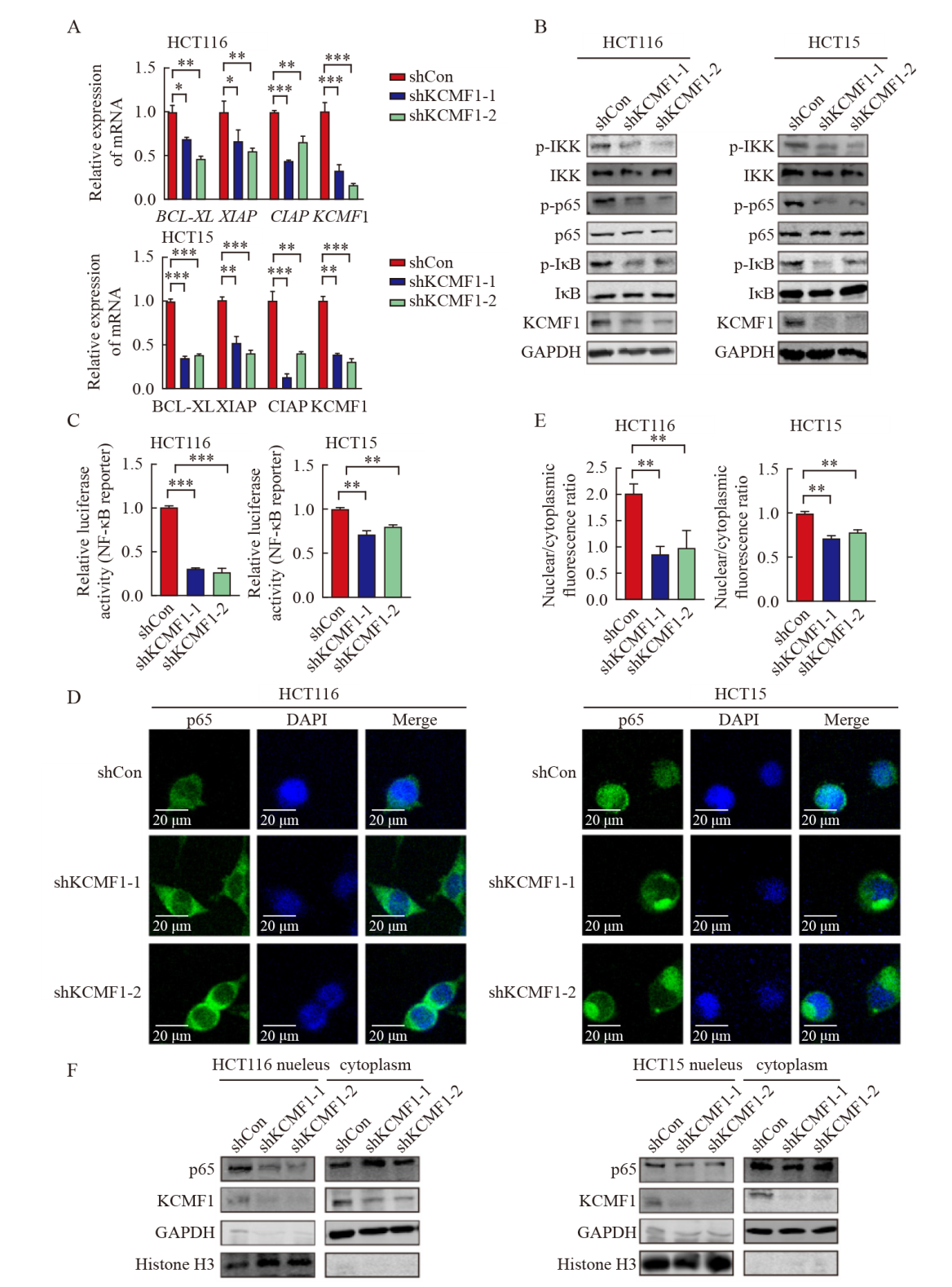
Knockdown of KCMF1 inhibits the activation of the NF-κB signaling in human CRC cells A: mRNA levels of KCMF1, BCL-XL, XIAP and CIAP in HCT116 and HCT15 cells with or without KCMF1 knockdown. *: P<0.05, assessed by Student-Newman-Keuls; **: P<0.01, assessed by Student-Newman-Keuls; ***: P<0.001, assessed by Student-Newman-Keuls. B: Expression of the phosphorylated proteins related to NF-κB signaling in HCT116 and HCT15 cells with or without KCMF1 knockdown. C: Activity of the NF-κB-reporter in HCT116 and HCT15 cells with KCMF1 knockdown. **: P<0.01, assessed by Student-Newman-Keuls; ***: P<0.001, assessed by Student-Newman-Keuls. D: Immunofluorescence of p65 (green) in HCT116 and HCT15 cells treated with TNFα (10 ng/mL) for 5 min after KCMF1 knockdown. DAPI stains nuclear DNA (blue). Scale=20 microns. E: Quantification of p65 nuclear and cytosolic localization was assessed in the indicated cells, as shown by a ratio of nuclear to cytoplasmic fluorescence. **: P<0.01, assessed by Student-Newman-Keuls. F: p65 cellular localization in HCT116 and HCT15 cells with or without KCMF1 knockdown."
| [1] | SUNG H, FERLAY J, SIEGEL R L, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71(3): 209-249. |
| [2] |
GUINNEY J, DIENSTMANN R, WANG X, et al. The consensus molecular subtypes of colorectal cancer[J]. Nat Med, 2015, 21(11): 1350-1356.
doi: 10.1038/nm.3967 pmid: 26457759 |
| [3] | LAURENT-PUIG P, AGOSTINI J, MALEY K. Colorectal oncogenesis[J]. Bull Cancer, 2010, 97(11): 1311-1321. |
| [4] | TAIXIANG W, MUNRO A J, GUANJIAN L. Chinese medical herbs for chemotherapy side effects in colorectal cancer patients[J]. Cochrane Database Syst Rev, 2005, 2005(1): CD004540. |
| [5] | RONNEKLEIV-KELLY S M, KENNEDY G D. Management of stage Ⅳ rectal cancer: palliative options[J]. World J Gastroenterol, 2011, 17(7): 835-847. |
| [6] | XIE Y H, CHEN Y X, FANG J Y. Comprehensive review of targeted therapy for colorectal cancer[J]. Signal Transduct Target Ther, 2020, 5(1): 22. |
| [7] |
GIANNAKIS M, MU X J, SHUKLA S A, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma[J]. Cell Rep, 2016, 17(4): 1206.
doi: S2211-1247(16)31395-X pmid: 27760322 |
| [8] |
HERSHKO A, CIECHANOVER A. The ubiquitin system[J]. Annu Rev Biochem, 1998, 67: 425-479.
pmid: 9759494 |
| [9] |
SWATEK K N, KOMANDER D. Ubiquitin modifications[J]. Cell Res, 2016, 26(4): 399-422.
doi: 10.1038/cr.2016.39 pmid: 27012465 |
| [10] | QI J F, RONAI Z A. Dysregulation of ubiquitin ligases in cancer[J]. Drug Resist Updat, 2015, 23: 1-11. |
| [11] |
BEILKE S, OSWALD F, GENZE F, et al. The zinc-finger protein KCMF1 is overexpressed during pancreatic cancer development and downregulation of KCMF1 inhibits pancreatic cancer development in mice[J]. Oncogene, 2010, 29(28): 4058-4067.
doi: 10.1038/onc.2010.156 pmid: 20473331 |
| [12] |
HONG J H, KAUSTOV L, COYAUD E, et al. KCMF1 (potassium channel modulatory factor 1) Links RAD6 to UBR4 (ubiquitin N-recognin domain-containing E3 ligase 4) and lysosome-mediated degradation[J]. Mol Cell Proteomics, 2015, 14(3): 674-685.
doi: 10.1074/mcp.M114.042168 pmid: 25582440 |
| [13] | HEO A J, KIM S B, JI C H, et al. The N-terminal cysteine is a dual sensor of oxygen and oxidative stress[J]. Proc Natl Acad Sci U S A, 2021, 118(50): e2107993118. |
| [14] | CERVIA L D, SHIBUE T, BORAH A A, et al. A ubiquitination cascade regulating the integrated stress response and survival in carcinomas[J]. Cancer Discov, 2023, 13(3): 766-795. |
| [15] | SINGH A, CHOUDHURY S D, SINGH P, et al. Disruption in networking of KCMF1 linked ubiquitin ligase impairs autophagy in CD8+ memory T cells of patients with renal cell carcinoma[J]. Cancer Lett, 2023, 564: 216194. |
| [16] | JANG J H. FIGC, a novel FGF-induced ubiquitin-protein ligase in gastric cancers[J]. FEBS Lett, 2004, 578(1/2): 21-25. |
| [17] | XI Y, XU P F. Global colorectal cancer burden in 2020 and projections to 2040[J]. Transl Oncol, 2021, 14(10): 101174. |
| [18] | YANG Y, WANG H Y, CHEN Y K, et al. Current status of surgical treatment of rectal cancer in China[J]. Chin Med J, 2020, 133(22): 2703-2711. |
| [19] |
VARLAND S, SILVA R D, KJOSÅS I, et al. N-terminal acetylation shields proteins from degradation and promotes age-dependent motility and longevity[J]. Nat Commun, 2023, 14(1): 6774.
doi: 10.1038/s41467-023-42342-y pmid: 37891180 |
| [20] | ZOU J, MI L, YU X F, et al. Interaction of 14-3-3σ with KCMF1 suppresses the proliferation and colony formation of human colon cancer stem cells[J]. World J Gastroenterol, 2013, 19(24): 3770-3780. |
| [21] |
SAKAMOTO K, MAEDA S, HIKIBA Y, et al. Constitutive NF-kappaB activation in colorectal carcinoma plays a key role in angiogenesis, promoting tumor growth[J]. Clin Cancer Res, 2009, 15(7): 2248-2258.
doi: 10.1158/1078-0432.CCR-08-1383 pmid: 19276252 |
| [22] |
RAJITHA B, BELALCAZAR A, NAGARAJU G P, et al. Inhibition of NF-κB translocation by curcumin analogs induces G0/G1 arrest and downregulates thymidylate synthase in colorectal cancer[J]. Cancer Lett, 2016, 373(2): 227-233.
doi: 10.1016/j.canlet.2016.01.052 pmid: 26850372 |
| [23] |
JANI T S, DEVECCHIO J, MAZUMDAR T, et al. Inhibition of NF-kappaB signaling by quinacrine is cytotoxic to human colon carcinoma cell lines and is synergistic in combination with tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) or oxaliplatin[J]. J Biol Chem, 2010, 285(25): 19162-19172.
doi: 10.1074/jbc.M109.091645 pmid: 20424169 |
| [24] |
SENFT D, QI J F, RONAI Z A. Ubiquitin ligases in oncogenic transformation and cancer therapy[J]. Nat Rev Cancer, 2018, 18(2): 69-88.
doi: 10.1038/nrc.2017.105 pmid: 29242641 |
| [25] |
HOU Y Z, MOREAU F, CHADEE K. PPARγ is an E3 ligase that induces the degradation of NFκB/p65[J]. Nat Commun, 2012, 3: 1300.
doi: 10.1038/ncomms2270 pmid: 23250430 |
| [26] | JI J X, DING K K, LUO T, et al. TRIM22 activates NF-κB signaling in glioblastoma by accelerating the degradation of IκBα[J]. Cell Death Differ, 2021, 28(1): 367-381. |
| [27] |
DENG L, WANG C, SPENCER E, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain[J]. Cell, 2000, 103(2): 351-361.
doi: 10.1016/s0092-8674(00)00126-4 pmid: 11057907 |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
沪ICP备12009617
Powered by Beijing Magtech Co. Ltd
