Welcome to China Oncology,
China Oncology ›› 2022, Vol. 32 ›› Issue (12): 1199-1209.doi: 10.19401/j.cnki.1007-3639.2022.12.008
• Article • Previous Articles Next Articles
CUI Zhongze( ), HE Shuang, WEN Feifei, LI Yangyang, XU Xiaoyang, LU Lizhen, WU Shuhua(
), HE Shuang, WEN Feifei, LI Yangyang, XU Xiaoyang, LU Lizhen, WU Shuhua( )
)
Received:2022-04-22
Revised:2022-10-17
Online:2022-12-30
Published:2023-02-02
Contact:
WU Shuhua
Share article
CLC Number:
CUI Zhongze, HE Shuang, WEN Feifei, LI Yangyang, XU Xiaoyang, LU Lizhen, WU Shuhua. Experimental study on influence of autophagy on DPD expression and its effect on chemotherapy with 5-FU in colorectal cancer[J]. China Oncology, 2022, 32(12): 1199-1209.
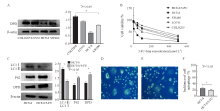
Fig.1
Levels of autophagy, DPD and drug resistance in colon cancer cells A: Expression level of DPD in different colon cancer cell lines; B: Sensitivity of different cell lines to 5-FU (dose-response curve); C: Expression of LC3, P62 and DPD in HCT-8 and HCT-8/5-FU cells; D: MDC method was used to detect the autophagy level of HCT-8 cells. E: MDC method was used to detect the autophagy level of HCT-8/5-FU cells. F: Incidence of autophagy in HCT-8 and HCT-8/5-FU cells."
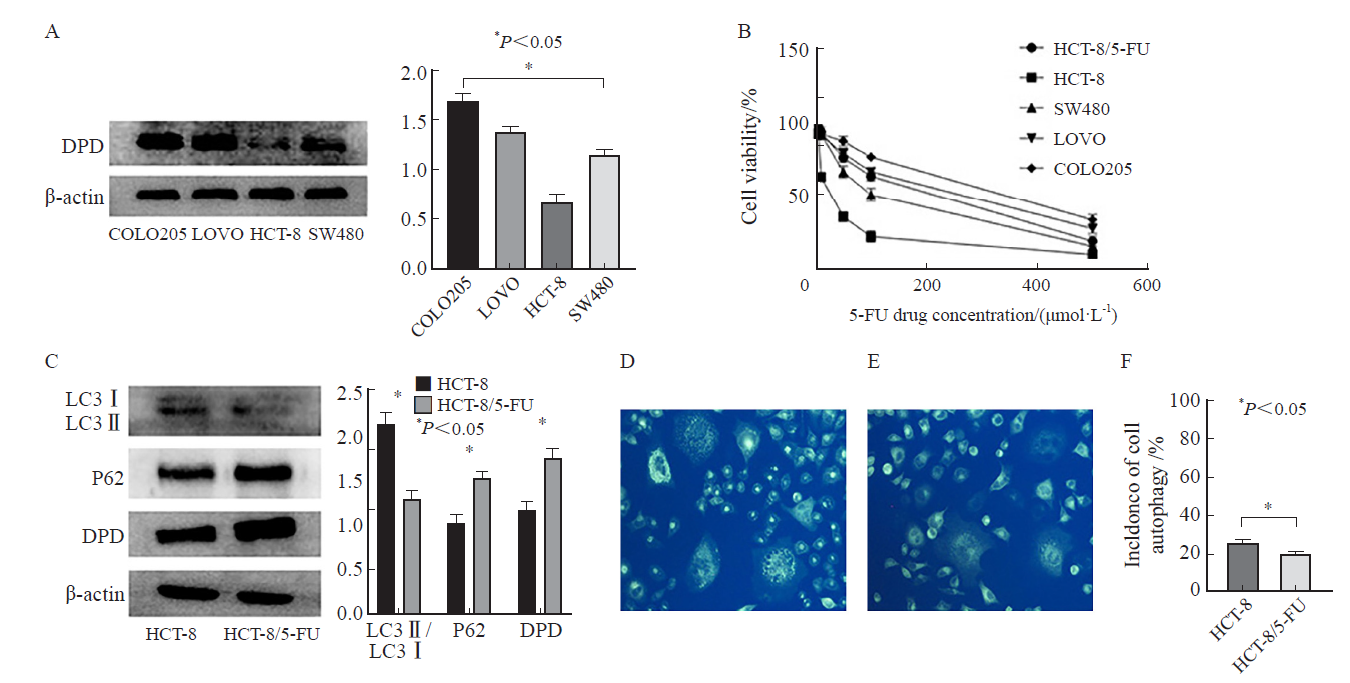

Fig.2
MDC staining to detect autophagy levels in different intervention groups A: HCT-8/5-FU cell control group; B: HCT-8/5-FU cell rapamycin intervention group; C: HCT-8/5-FU cells 3-MA intervention group; D: HCT-8/5-FU cells hydroxychloroquine intervention group; E: Autophagy expression rate of HCT-8/5-FU cells in each group; F: HCT-8 cell control group; G: HCT-8 cell rapamycin intervention group; H: HCT-8 cells 3-MA intervention group; I: HCT-8 cell hydroxychloroquine intervention group; J: Autophagy expression rate of HCT-8 cells in each group."
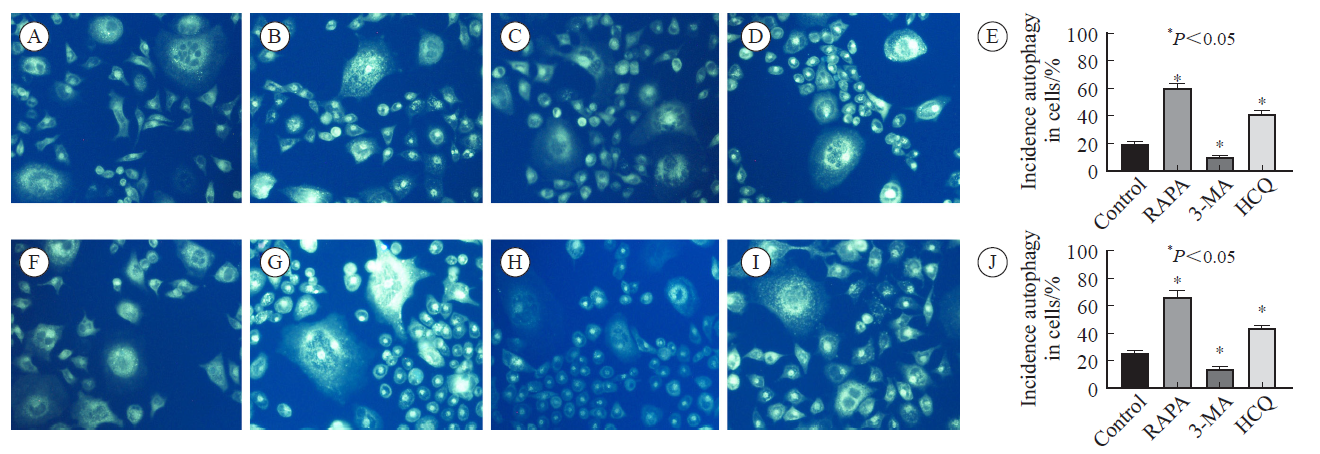
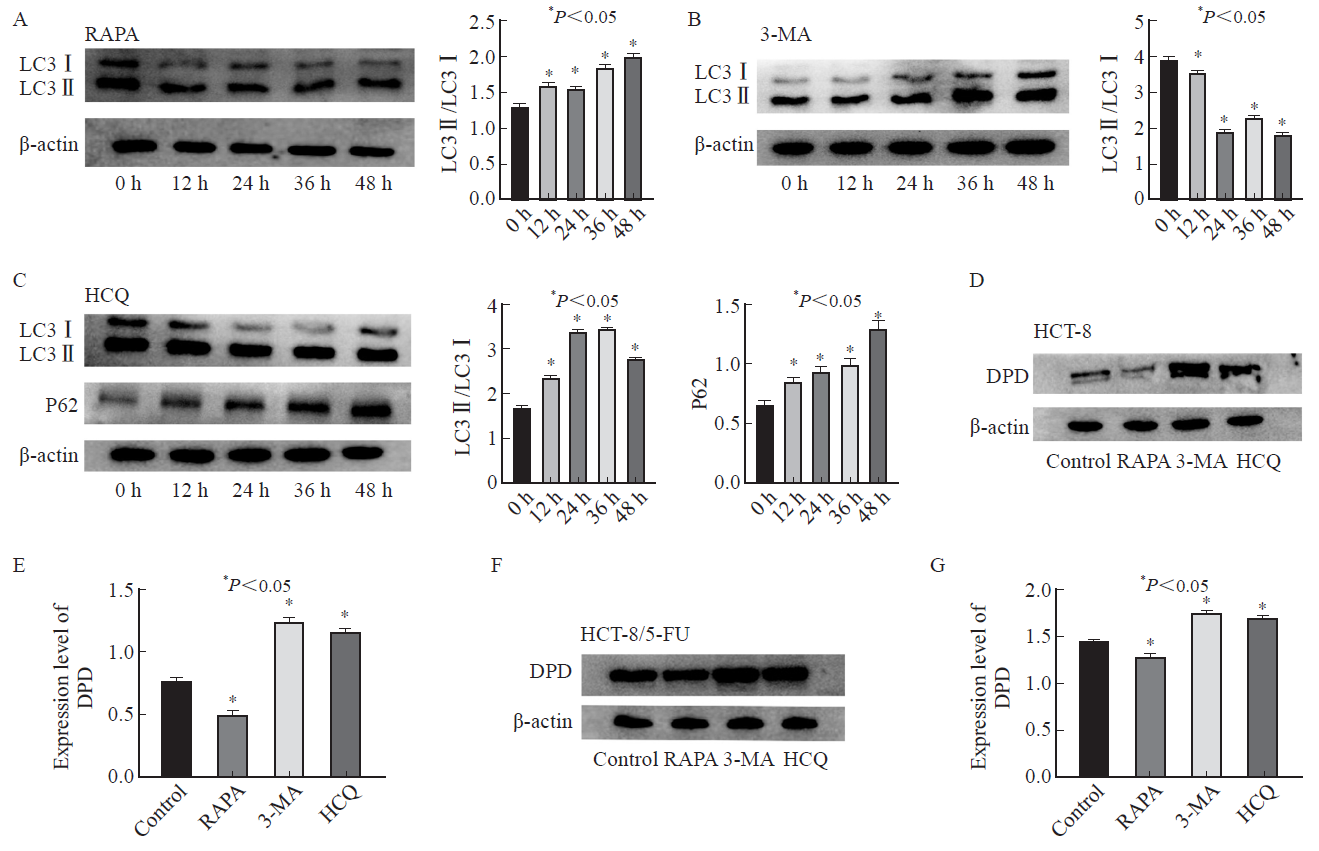
Fig. 3
Changes of autophagy-related proteins and DPD in cells after intervention with autophagy inhibitors and activators A: HCT-8/5-FU cell rapamycin intervention group; B: HCT-8/5-FU cells 3-MA intervention group; C: HCT-8/5-FU cells hydroxychloroquine intervention group; D-E: DPD content after HCT-8 cell intervention; F-G: DPD content after intervention with HCT-8/5-FU cells.RAPA: Rapamycin."
Tab. 1
CCK-8 assay was used to detect the survival rate of HCT-8 and HCT-8/5-FU cell lines after different drug intervention ($\bar{x}\pm s$)"
| Variable | Control | Rapamycin | 3-MA | HCQ |
|---|---|---|---|---|
| HCT-8 | ||||
| Autophagy intervention | 100±1.91 | 93.41±0.72 | 92.33±1.11 | 91.21±1.28 |
| Combined application of 5-FU and autophagy intervention | 50.00±0.95 | 22.92±2.36* | 45.67±1.98 | 44.42±3.04 |
| HCT-8/5-FU | ||||
| Autophagy intervention | 100±1.36 | 93.25±1.32 | 92.67±0.90 | 91.57±0.95 |
| Combined application of 5-FU and autophagy intervention | 50.00±1.04 | 26.26±1.93* | 46.56±2.26 | 45.72±2.05 |
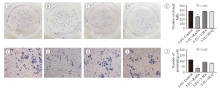
Fig.4
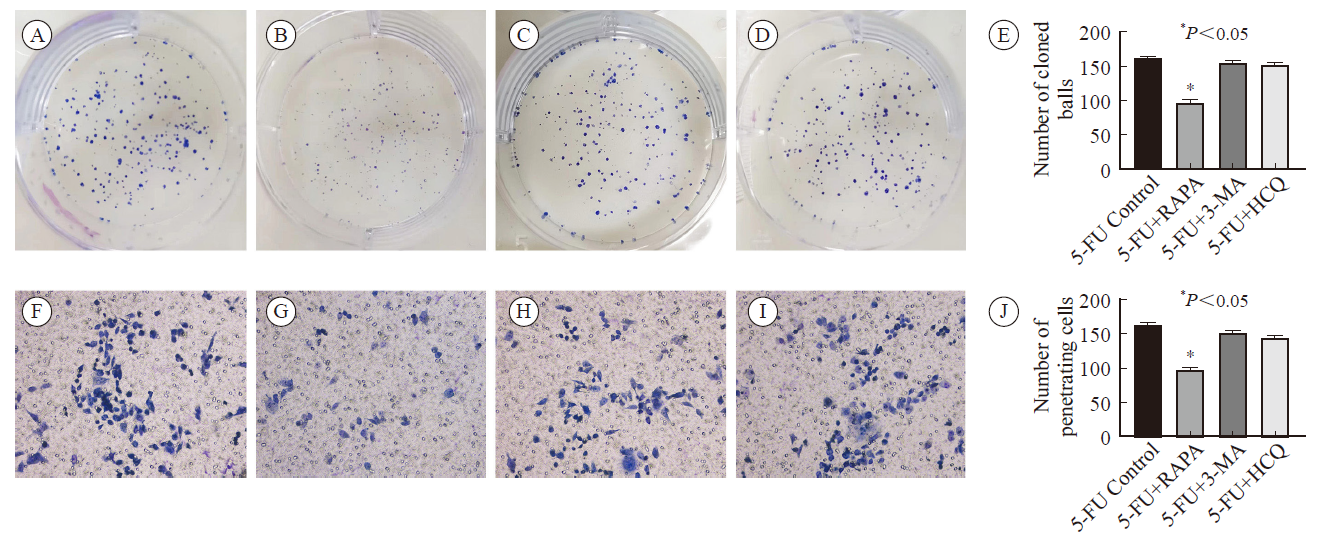
Plate cloning assay (500 cells/hole) and transwell assay were used to detect the proliferation and invasion ability of HCT-8 cell lines after different interventions A-E: Colony-forming assay. A: 5-FU control group; B: 5-FU + rapamycin group; C: 5-FU + 3-MA group; D: 5-FU + hydroxychloroquine group; E: Incidence of autophagy in HCT-8 cell lines after different interventions; F-I: Cell invasion assay: F: 5-FU control group; G: 5-FU + rapamycin group; H: 5-FU + 3-MA group; I: 5-FU + hydroxychloroquine group; J: Penetration cell number of HCT-8 cell line after different interventions."
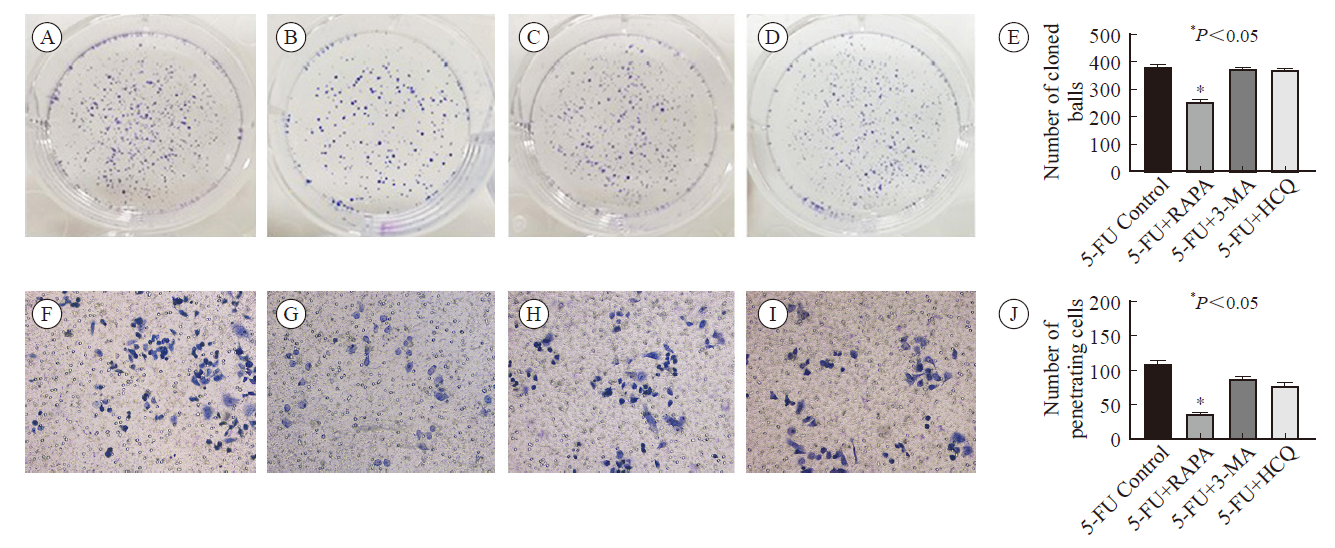
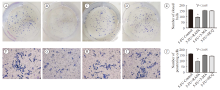
Fig. 5
Plate cloning assay (200 cells/hole) and transwell assay were used to detect the proliferation and invasion ability of HCT-8/5-FU cell lines after different interventions Colony-forming assay: A: 5-FU control group; B: 5-FU + rapamycin group; C: 5-FU + 3-MA group; D: 5-FU + hydroxychloroquine group; E: Incidence of autophagy in HCT-8/5-FU cell lines after different interventions Cell invasion assay: F: 5-FU control group; G: 5-FU + rapamycin group; H: 5-FU + 3-MA group; I: 5-FU + hydroxychloroquine group; J: Penetration cell number of HCT-8/FU cell line after different interventions"
| [1] |
PATEL S G, KARLITZ J J, YEN T, et al. The rising tide of early-onset colorectal cancer: a comprehensive review of epidemiology, clinical features, biology, risk factors, prevention, and early detection[J]. Lancet Gastroenterol Hepatol, 2022, 7(3): 262-274.
doi: 10.1016/S2468-1253(21)00426-X |
| [2] |
BENSON A B, VENOOK A P, AL-HAWARY M M, et al. NCCN guidelines insights: colon cancer, version 2.2018[J]. J Natl Compr Canc Netw, 2018, 16(4): 359-369.
doi: 10.6004/jnccn.2018.0021 |
| [3] |
MALIER M, GHARZEDDINE K, LAVERRIERE M H, et al. Hypoxia drives dihydropyrimidine dehydrogenase expression in macrophages and confers chemoresistance in colorectal cancer[J]. Cancer Res, 2021, 81(23): 5963-5976.
doi: 10.1158/0008-5472.CAN-21-1572 pmid: 34645611 |
| [4] | DE FALCO V, NATALICCHIO M I, NAPOLITANO S, et al. A case report of a severe fluoropyrimidine-related toxicity due to an uncommon DPYD variant[J]. Medicine, 2019, 98(21): e15759. |
| [5] | DEAN L, KANE M. Fluorouracil therapy and DPYD genotype[M]. PRATT V M, SCOTT S A, PIRMOHAMED M, eds. Medical genetics summaries. . Bethesda (MD): National Center for Biotechnology Information (US); November 3, 2016. |
| [6] |
贾真真, 何双, 李扬扬, 等. 结直肠癌中DPD与LC3、P62表达的相关性及其临床意义[J]. 中国癌症杂志, 2022, 32(1): 24-33.
doi: 10.19401/j.cnki.1007-3639.2022.01.003 |
| JIA Z Z, HE S, LI Y Y, et al. Correlations between expressions of DPD, LC3 and P62 in colorectal cancer and their clinical significance[J]. China Oncol, 2022, 32(1): 24-33. | |
| [7] |
YU L, CHEN Y, TOOZE S A. Autophagy pathway: cellular and molecular mechanisms[J]. Autophagy, 2018, 14(2): 207-215.
doi: 10.1080/15548627.2017.1378838 pmid: 28933638 |
| [8] |
DIKIC I, ELAZAR Z. Mechanism and medical implications of mammalian autophagy[J]. Nat Rev Mol Cell Biol, 2018, 19(6): 349-364.
doi: 10.1038/s41580-018-0003-4 |
| [9] |
ISLAM M A, SOORO M A, ZHANG P H. Autophagic regulation of p62 is critical for cancer therapy[J]. Int J Mol Sci, 2018, 19(5): 1405.
doi: 10.3390/ijms19051405 |
| [10] |
JIN H, SEO G S, LEE S H. Isoliquiritigenin-mediated p62/SQSTM1 induction regulates apoptotic potential through attenuation of caspase-8 activation in colorectal cancer cells[J]. Eur J Pharmacol, 2018, 841: 90-97.
doi: S0014-2999(18)30591-0 pmid: 30339814 |
| [11] |
LAMARK T, SVENNING S, JOHANSEN T. Regulation of selective autophagy: the p62/SQSTM1 paradigm[J]. Essays Biochem, 2017, 61(6): 609-624.
doi: 10.1042/EBC20170035 pmid: 29233872 |
| [12] |
SUN A Q, WEI J, CHILDRESS C, et al. The E3 ubiquitin ligase NEDD4 is an LC3-interactive protein and regulates autophagy[J]. Autophagy, 2017, 13(3): 522-537.
doi: 10.1080/15548627.2016.1268301 pmid: 28085563 |
| [13] |
LEVINE B, KROEMER G. Biological functions of autophagy genes: a disease perspective[J]. Cell, 2019, 176(1/2): 11-42.
doi: 10.1016/j.cell.2018.09.048 |
| [14] |
BONAM S R, TRANCHANT C, MULLER S. Autophagy-lysosomal pathway as potential therapeutic target in Parkinson's disease[J]. Cells, 2021, 10(12): 3547.
doi: 10.3390/cells10123547 |
| [15] |
YANG Y, WANG Q, SONG D J, et al. Lysosomal dysfunction and autophagy blockade contribute to autophagy-related cancer suppressing peptide-induced cytotoxic death of cervical cancer cells through the AMPK/mTOR pathway[J]. J Exp Clin Cancer Res, 2020, 39(1): 197.
doi: 10.1186/s13046-020-01701-z |
| [16] | LI J X, LIU G, LI L, et al. Research progress on the effect of autophagy-lysosomal pathway on tumor drug resistance[J]. Exp Cell Res, 2020, 389(2): 111925. |
| [17] |
XUAN Y, ZHAO S, XIAO X J, et al. Inhibition of chaperone-mediated autophagy reduces tumor growth and metastasis and promotes drug sensitivity in colorectal cancer[J]. Mol Med Rep, 2021, 23(5): 360.
doi: 10.3892/mmr.2021.11999 |
| [18] | YANG J W, ZHANG Q H, LIU T. Autophagy facilitates anticancer effect of 5-fluorouracil in HCT-116 cells[J]. J Cancer Res Ther, 2018, 14(Supplement): S1141-S1147. |
| [19] |
TOZER T, HEALE K, MANTO CHAGAS C, et al. Interdomain twists of human thymidine phosphorylase and its active-inactive conformations: binding of 5-FU and its analogues to human thymidine phosphorylase versus dihydropyrimidine dehydrogenase[J]. Chem Biol Drug Des, 2019, 94(5): 1956-1972.
doi: 10.1111/cbdd.13596 pmid: 31356728 |
| [20] |
SHARMA V, GUPTA S K, VERMA M. Dihydropyrimidine dehydrogenase in the metabolism of the anticancer drugs[J]. Cancer Chemother Pharmacol, 2019, 84(6): 1157-1166.
doi: 10.1007/s00280-019-03936-w |
| [21] |
ZHANG Y H, SHI W N, WU S H, et al. SphK2 confers 5-fluorouracil resistance to colorectal cancer via upregulating H3K56ac-mediated DPD expression[J]. Oncogene, 2020, 39(29): 5214-5227.
doi: 10.1038/s41388-020-1352-y |
| [22] | ZHANG Y H, LUO D D, WAN S B, et al. S1PR2 inhibitors potently reverse 5-FU resistance by downregulating DPD expression in colorectal cancer[J]. Pharmacol Res, 2020, 155: 104717. |
| [23] |
YOSHII S R, MIZUSHIMA N. Monitoring and measuring autophagy[J]. Int J Mol Sci, 2017, 18(9): 1865.
doi: 10.3390/ijms18091865 |
| [24] | OZATES N P, SOĞUTLU F, LERMINOGLU F, et al. Effects of rapamycin and AZD3463 combination on apoptosis, autophagy, and cell cycle for resistance control in breast cancer[J]. Life Sci, 2021, 264: 118643. |
| [25] |
ZHANG J, MAO W, LIU Y Y, et al. 3-MA enhanced chemosensitivity in cisplatin resistant hypopharyngeal squamous carcinoma cells via inhibiting beclin-1 mediated autophagy[J]. Curr Pharm Des, 2021, 27(7): 996-1005.
doi: 10.2174/1381612826666201221150431 |
| [26] | LIU L Q, WANG S B, SHAO Y F, et al. Hydroxychloroquine potentiates the anti-cancer effect of bevacizumab on glioblastoma via the inhibition of autophagy[J]. Biomedecine Pharmacother, 2019, 118: 109339. |
| [27] | ZAMAME RAMIREZ J A, ROMAGNOLI G G, KANENO R. Inhibiting autophagy to prevent drug resistance and improve anti-tumor therapy[J]. Life Sci, 2021, 265: 118745. |
| [28] |
LI J, HOU N, FARIED A, et al. Inhibition of autophagy augments 5-fluorouracil chemotherapy in human colon cancer in vitro and in vivo model[J]. Eur J Cancer, 2010, 46(10): 1900-1909.
doi: 10.1016/j.ejca.2010.02.021 pmid: 20231086 |
| [29] |
CHOI J H, YOON J S, WON Y W, et al. Chloroquine enhances the chemotherapeutic activity of 5-fluorouracil in a colon cancer cell line via cell cycle alteration[J]. APMIS, 2012, 120(7): 597-604.
doi: 10.1111/j.1600-0463.2012.02876.x |
| [30] |
LIU W J, YE L, HUANG W F, et al. p62 links the autophagy pathway and the ubiqutin-proteasome system upon ubiquitinated protein degradation[J]. Cell Mol Biol Lett, 2016, 21: 29.
doi: 10.1186/s11658-016-0031-z pmid: 28536631 |
| [31] |
JOHANSEN T, LAMARK T. Selective autophagy mediated by autophagic adapter proteins[J]. Autophagy, 2011, 7(3): 279-296.
doi: 10.4161/auto.7.3.14487 pmid: 21189453 |
| [32] |
ZHENG N, SHABEK N. Ubiquitin ligases: Structure, function, and regulation[J]. Annu Rev Biochem, 2017, 86: 129-157.
doi: 10.1146/annurev-biochem-060815-014922 pmid: 28375744 |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
沪ICP备12009617
Powered by Beijing Magtech Co. Ltd
