Welcome to China Oncology,
China Oncology ›› 2024, Vol. 34 ›› Issue (6): 537-547.doi: 10.19401/j.cnki.1007-3639.2024.06.002
• Article • Previous Articles Next Articles
DONG Jianqiao1( ), LI Kunyan1, LI Jing2, WANG Bin2, WANG Yanhong3, JIA Hongyan2(
), LI Kunyan1, LI Jing2, WANG Bin2, WANG Yanhong3, JIA Hongyan2( )(
)( )
)
Received:2024-03-07
Revised:2024-06-07
Online:2024-06-30
Published:2024-07-16
Share article
CLC Number:
DONG Jianqiao, LI Kunyan, LI Jing, WANG Bin, WANG Yanhong, JIA Hongyan. A study on mechanism of SIRT3 inducing endocrine drug resistance in breast cancer via deacetylating YME1L1[J]. China Oncology, 2024, 34(6): 537-547.
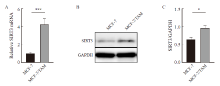
Fig. 2
SIRT3 is highly expressed in tamoxifen-resistant breast cancer. A: SIRT3 mRNA expression was detected by RTFQ-PCR. B, C: SIRT3 protein expression and quantification in both cell lines were detected by Western blot. *: P<0.05, compared with each other; ***: P<0.001, compared with each other."
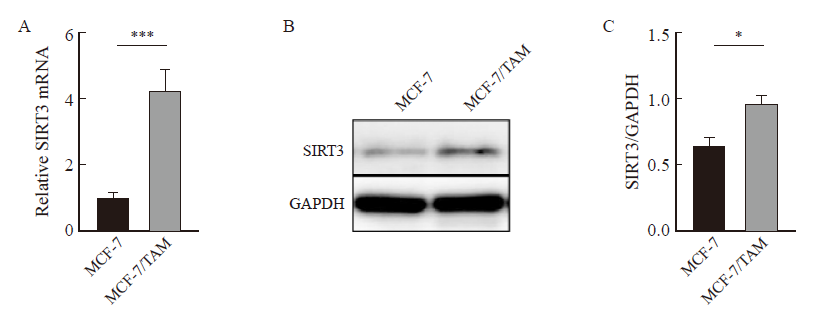
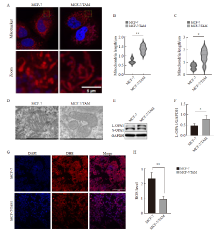
Fig. 3
Detection of mitochondrial morphology and function in breast cancer after endocrine resistance A, B: Mitochondrial length was observed and quantified by fluorescence staining (scale bar=5 μm); C, D: Mitochondrial morphology was observed and quantified by transmission electron microscopy (scale bar=200 nm); E, F: Western blot was used to detect and quantify L-OPA1 protein in parental cells and drug-resistant cells. G, H: DHE staining assessment, ROS levels and quantification in parental and resistant cells (scale bar=200 μm). *: P<0.05, compared with each other; **: P<0.01, compared with each other."
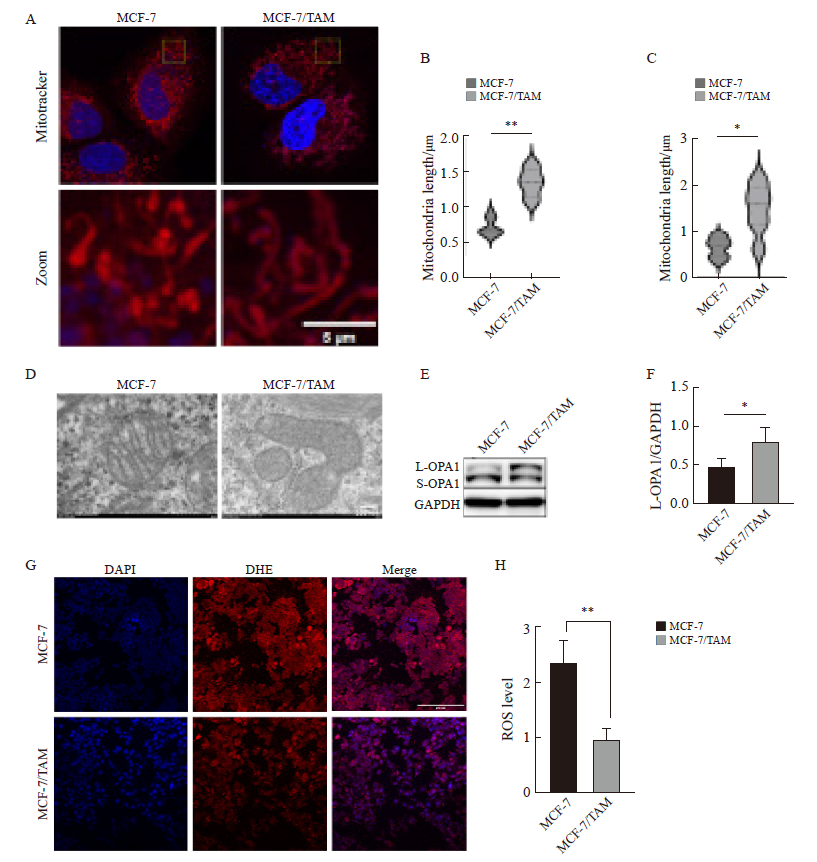
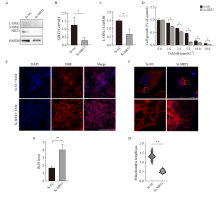
Fig. 4
Cell proliferation, mitochondrial morphology, and function were examined by SIRT3 knockdown in drug-resistant cells A, B, C: Western blot was used to detect the expression and quantification of SIRT3 protein and L-OPA1 protein after SIRT3 knockdown. D: CCK-8 assay was used to detect the cell viability under tamoxifen treatment; E, G: DHE probe was used to detect the intracellular ROS level and quantification after SIRT3 knockdown (scale bar=200 μm); F, H: Immunofluorescence detection of mitochondrial length changes and quantification after SIRT3 knockdown (scale bar=5 μm). *: P<0.05, Si-NC compared with Si-SIRT3."
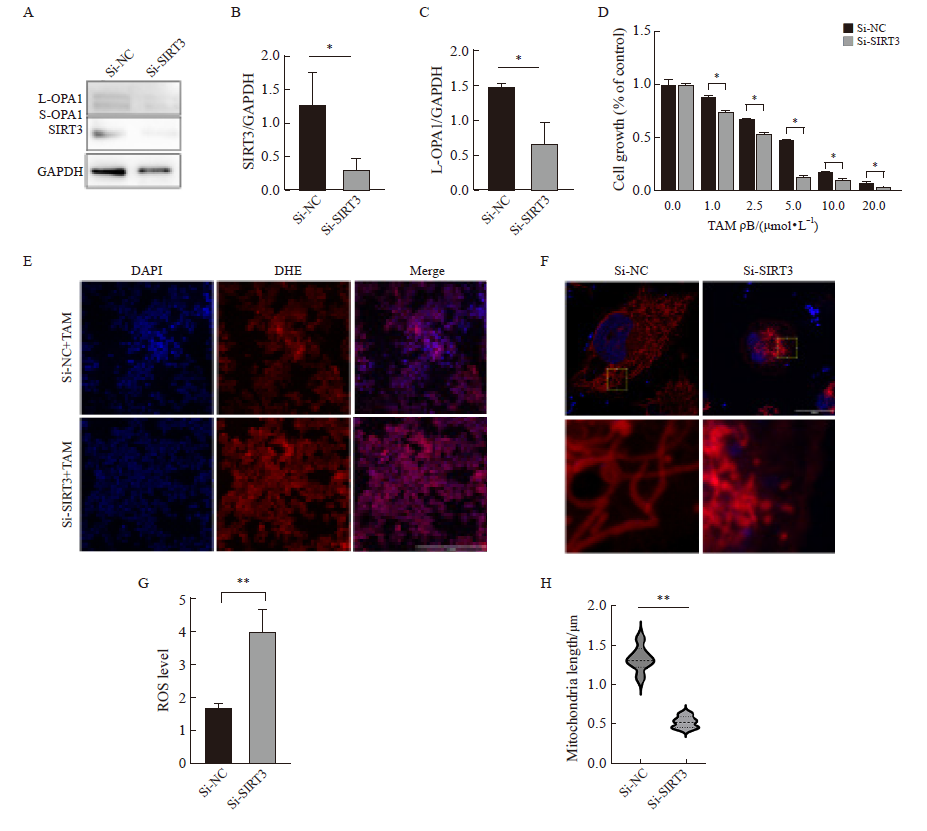

Fig. 5
Overexpression of SIRT3 in parental cells examined cell proliferation, mitochondrial morphology, and function A, B, C: Western blot was used to detect the expression and quantification of L-OPA1 protein after SIRT3 overexpression; D: After SIRT3 was overexpressed in parental cells, CCK-8 assay was used to detect the cell viability under tamoxifen treatment; E, G: Detection and quantification of mitochondrial membrane potential by JC-1 probe (scale bar=200 μm); H, I: Fluorescence staining for mitochondrial morphology and quantification (scale bar=5 μm). *: P<0.05, compared with each other; **: P<0.01, compared with each other; ***: P<0.001, compared with OE-SIRT3 group."
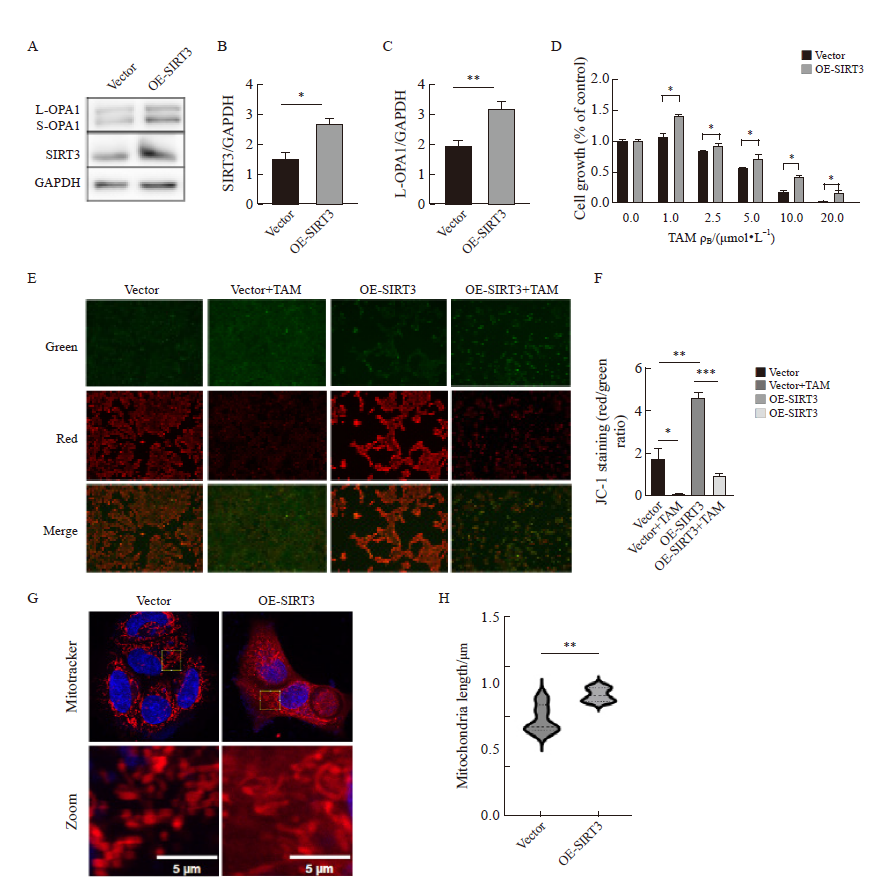
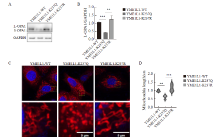
Fig. 6
Changes in mitochondrial morphology caused by overexpression of YME1L1 in parental cells A, B: YME1L1 was overexpressed, and the expression and quantification of L-OPA1 protein were detected by Western blot. C, D: Length and quantification of mitochondria by fluorescent staining (scale bar=5 μm). **: P<0.01, compared with each other; ***: P<0.001, compared with YME1L1-K237Q."
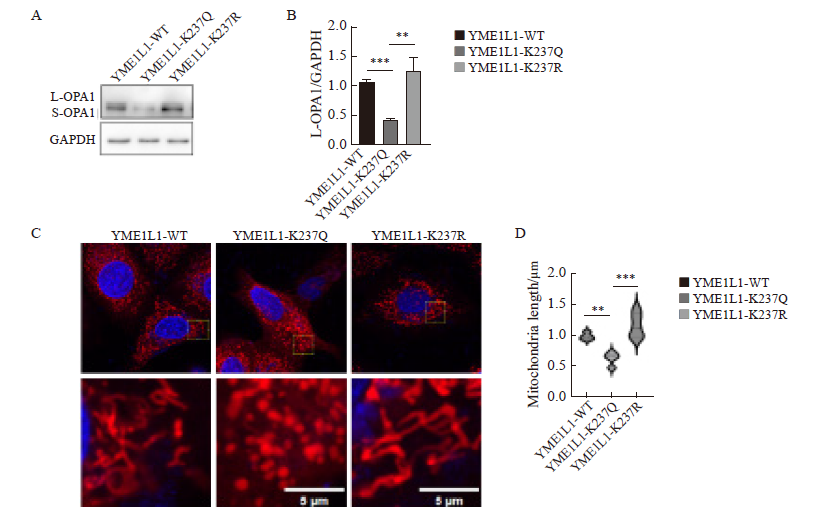

Fig. 7
SIRT3 regulates the acetylation status of YME1L1 A-G: IP was used to detect the interaction between YME1L1 and SIRT3 and quantify. *: P<0.05, compared with each other; **: P<0.01, compared with each other; ***: P<0.001, compared with each other. H: Co-localization of YME1L1 and SIRT3 was detected by immunofluorescence (scale bar=50 μm)."
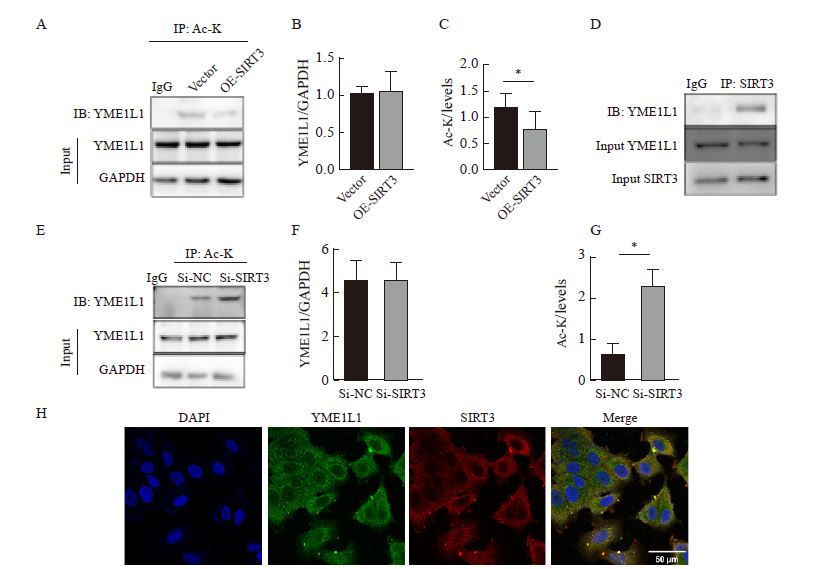
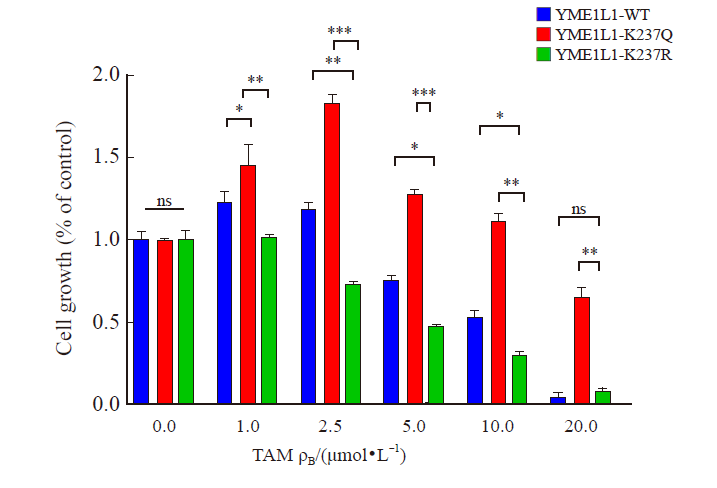
Fig. 8
Deacetylating Y ME1L1 promotes tamoxifen resistance in breast cancer CCK-8 assay was used to detect cell viability under tamoxifen treatment. *: P<0.05, compared with each other; **: P<0.01, compared with each other; ***: P<0.001, compared with each other. ns: No significance, compared with YME1L1-WT or YME1L1-K237R."
| [1] | BRAY F, LAVERSANNE M, SUNG H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA A Cancer J Clin, 2024, 74(3): 229-263. |
| [2] | CAO W, CHEN H D, YU Y W, et al. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020[J]. Chin Med J, 2021, 134(7): 783-791. |
| [3] | SIEGEL R L, GIAQUINTO A N, JEMAL A. Cancer statistics, 2024[J]. CA Cancer J Clin, 2024, 74(1): 12-49. |
| [4] | VOUDOURI K, BERDIAKI A, TZARDI M, et al. Insulin-like growth factor and epidermal growth factor signaling in breast cancer cell growth: focus on endocrine resistant disease[J]. Anal Cell Pathol, 2015, 2015: 975495. |
| [5] | ZAHEDIPOUR F, JAMIALAHMADI K, KARIMI G. The role of noncoding RNAs and sirtuins in cancer drug resistance[J]. Eur J Pharmacol, 2020, 877: 173094. |
| [6] | LIU F, YUAN L H, LI L, et al. S-sulfhydration of SIRT3 combats BMSC senescence and ameliorates osteoporosis via stabilizing heterochromatic and mitochondrial homeostasis[J]. Pharmacol Res, 2023, 192: 106788. |
| [7] |
ZHANG J, XIANG H G, LIU J, et al. Mitochondrial Sirtuin 3: new emerging biological function and therapeutic target[J]. Theranostics, 2020, 10(18): 8315-8342.
doi: 10.7150/thno.45922 pmid: 32724473 |
| [8] |
ZHANG L, REN X C, CHENG Y, et al. Identification of Sirtuin 3, a mitochondrial protein deacetylase, as a new contributor to tamoxifen resistance in breast cancer cells[J]. Biochem Pharmacol, 2013, 86(6): 726-733.
doi: 10.1016/j.bcp.2013.06.032 pmid: 23856293 |
| [9] | HUBER-KEENER K J, LIU X P, WANG Z, et al. Differential gene expression in tamoxifen-resistant breast cancer cells revealed by a new analytical model of RNA-Seq data[J]. PLoS One, 2012, 7(7): e41333. |
| [10] | Disturbance of mitochondrial dynamics and mitophagy in sepsis-induced acute kidney injury-PubMed[Internet]. [cited 2024 Mar 2]. https://pubmed.ncbi.nlm.nih.gov/31479679/. |
| [11] |
BAEK M L, LEE J, PENDLETON K E, et al. Mitochondrial structure and function adaptation in residual triple negative breast cancer cells surviving chemotherapy treatment[J]. Oncogene, 2023, 42(14): 1117-1131.
doi: 10.1038/s41388-023-02596-8 pmid: 36813854 |
| [12] | ADEBAYO M, SINGH S, SINGH A P, et al. Mitochondrial fusion and fission: the fine-tune balance for cellular homeostasis[J]. FASEB J, 2021, 35(6): e21620. |
| [13] | RUAN Y, LI H, ZHANG K, et al. Loss of Yme1L perturbates mitochondrial dynamics[J]. Cell Death Dis, 2013, 4(10): e896. |
| [14] | ZAMBERLAN M, BOECKX A, MULLER F, et al. Inhibition of the mitochondrial protein Opa1 curtails breast cancer growth[J]. J Exp Clin Cancer Res, 2022, 41(1): 95. |
| [15] |
HANKER A B, SUDHAN D R, ARTEAGA C L. Overcoming endocrine resistance in breast cancer[J]. Cancer Cell, 2020, 37(4): 496-513.
doi: S1535-6108(20)30149-5 pmid: 32289273 |
| [16] |
CARRICO C, MEYER J G, HE W J, et al. The mitochondrial acylome emerges: proteomics, regulation by sirtuins, and metabolic and disease implications[J]. Cell Metab, 2018, 27(3): 497-512.
doi: S1550-4131(18)30068-8 pmid: 29514063 |
| [17] | LIU L G, LI Y, CAO D Y, et al. SIRT3 inhibits gallbladder cancer by induction of AKT-dependent ferroptosis and blockade of epithelial-mesenchymal transition[J]. Cancer Lett, 2021, 510: 93-104. |
| [18] | S O, Q Z, L L, K Z, Z L, P L, et al. The double-edged sword of SIRT3 in cancer and its therapeutic applications[J]. Front Pharmacol, 2022, 13: 871560. |
| [19] | KENNY T C, HART P, RAGAZZI M, et al. Selected mitochondrial DNA landscapes activate the SIRT3 axis of the UPRmt to promote metastasis[J]. Oncogene, 2017, 36(31): 4393-4404. |
| [20] | ASHRAF N, ZINO S, MACINTYRE A, et al. Altered sirtuin expression is associated with node-positive breast cancer[J]. Br J Cancer, 2006, 95(8): 1056-1061. |
| [21] |
CHEN S Y, YANG X, YU M, et al. SIRT3 regulates cancer cell proliferation through deacetylation of PYCR1 in proline metabolism[J]. Neoplasia, 2019, 21(7): 665-675.
doi: S1476-5586(18)30666-3 pmid: 31108370 |
| [22] | KIM H S, PATEL K, MULDOON-JACOBS K, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress[J]. Cancer Cell, 2010, 17(1): 41-52. |
| [23] |
DESOUKI M M, DOUBINSKAIA I, GIUS D, et al. Decreased mitochondrial SIRT3 expression is a potential molecular biomarker associated with poor outcome in breast cancer[J]. Hum Pathol, 2014, 45(5): 1071-1077.
doi: 10.1016/j.humpath.2014.01.004 pmid: 24746213 |
| [24] |
FINLEY L W S, CARRACEDO A, LEE J, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization[J]. Cancer Cell, 2011, 19(3): 416-428.
doi: 10.1016/j.ccr.2011.02.014 pmid: 21397863 |
| [25] | ANDREWS R M, KUBACKA I, CHINNERY P F, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA[J]. Nat Genet, 1999, 23(2): 147. |
| [26] | OHBA Y, MACVICAR T, LANGER T. Regulation of mitochondrial plasticity by the i-AAA protease YME1L[J]. Biol Chem, 2020, 401(6/7): 877-890. |
| [27] |
ANAND R, WAI T, BAKER M J, et al. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission[J]. J Cell Biol, 2014, 204(6): 919-929.
doi: 10.1083/jcb.201308006 pmid: 24616225 |
| [28] |
HERKENNE S, EK O, ZAMBERLAN M, et al. Developmental and tumor angiogenesis requires the mitochondria-shaping protein Opa1[J]. Cell Metab, 2020, 31(5): 987-1003.e8.
doi: S1550-4131(20)30189-3 pmid: 32315597 |
| [1] | XU Rui, WANG Zehao, WU Jiong. Advances in the role of tumor-associated neutrophils in the development of breast cancer [J]. China Oncology, 2024, 34(9): 881-889. |
| [2] | CAO Xiaoshan, YANG Beibei, CONG Binbin, LIU Hong. The progress of treatment for brain metastases of triple-negative breast cancer [J]. China Oncology, 2024, 34(8): 777-784. |
| [3] | ZHANG Jian. Clinical consideration of two key questions in assessing menopausal status of female breast cancer patients [J]. China Oncology, 2024, 34(7): 619-627. |
| [4] | JIANG Dan, SONG Guoqing, WANG Xiaodan. Study on the mechanism of mitochondrial dysfunction and CPT1A/ERK signal transduction pathway regulating malignant behavior in breast cancer [J]. China Oncology, 2024, 34(7): 650-658. |
| [5] | HAO Xian, HUANG Jianjun, YANG Wenxiu, LIU Jinting, ZHANG Junhong, LUO Yubei, LI Qing, WANG Dahong, GAO Yuwei, TAN Fuyun, BO Li, ZHENG Yu, WANG Rong, FENG Jianglong, LI Jing, ZHAO Chunhua, DOU Xiaowei. Establishment of primary breast cancer cell line as new model for drug screening and basic research [J]. China Oncology, 2024, 34(6): 561-570. |
| [6] | Committee of Breast Cancer Society, China Anti-Cancer Association. Expert consensus on clinical applications of ovarian function suppression for Chinese women with early breast cancer (2024 edition) [J]. China Oncology, 2024, 34(3): 316-333. |
| [7] | ZHANG Qi, XIU Bingqiu, WU Jiong. Progress of important clinical research of breast cancer in China in 2023 [J]. China Oncology, 2024, 34(2): 135-142. |
| [8] | ZHANG Siyuan, JIANG Zefei. Important research progress in clinical practice for advanced breast cancer in 2023 [J]. China Oncology, 2024, 34(2): 143-150. |
| [9] | WANG Zhaobu, LI Xing, YU Xinmiao, JIN Feng. Important research progress in clinical practice for early breast cancer in 2023 [J]. China Oncology, 2024, 34(2): 151-160. |
| [10] | LUO Yang, SUN Tao, SHAO Zhimin, CUI Jiuwei, PAN Yueyin, ZHANG Qingyuan, CHENG Ying, LI huiping, YANG Yan, YE Changsheng, YU Guohua, WANG Jingfen, LIU Yunjiang, LIU Xinlan, ZHOU Yuhong, BAI Yuju, GU Yuanting, WANG Xiaojia, XU Binghe, SONG Lihua. Efficacy, metabolic characteristics, safety and immunogenicity of AK-HER2 compared with reference trastuzumab in patients with metastatic HER2-positive breast cancer: a multicenter, randomized, double-blind phase Ⅲ equivalence trial [J]. China Oncology, 2024, 34(2): 161-175. |
| [11] | CHEN Yuanxiang, YU Tao, YANG Shiyu, ZENG Tao, WEI Lan, ZHANG Yan. KDM4A promotes the migration and invasion of breast cancer cell line MDA-MB-231 by downregulating BMP9 [J]. China Oncology, 2024, 34(2): 176-184. |
| [12] | HU Xiaoyu, CAI Yuwen, YE Fugui, SHAO Zhimin, HU Weigang, YU Keda. Impact of BRCA1/2 germline mutation on the incidence of second primary cancer following postoperative radiotherapy in patients with triple-negative breast cancer [J]. China Oncology, 2024, 34(2): 185-190. |
| [13] | ZHANG Siwei, MA Ding, JIANG Yizhou, SHAO Zhimin. “Subtype-precise” therapy leads diagnostic and therapeutic innovations: a new pattern for precision treatment of breast cancer [J]. China Oncology, 2024, 34(11): 1045-1052. |
| [14] | OUYANG Fei, WANG Yang, CHEN Yu, PEI Guoqing, WANG Ling, ZHANG Yang, SHI Lei. Construction of the prediction model of breast cancer bone metastasis based on machine learning [J]. China Oncology, 2024, 34(10): 903-914. |
| [15] | ZHAO Junxiu, ZHU Yi, SONG Xiaoyu, ZHE Chao, XIAO Yuhan, LIU Yunduo, LI Linhai, XIAO Bin. Circ-0007766 acts as a miR-1972 sponge to promote breast cancer cell migration and invasion via upregulation of HER2 [J]. China Oncology, 2024, 34(10): 915-930. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
沪ICP备12009617
Powered by Beijing Magtech Co. Ltd
